Rep:Mod:ZZY666
Introduction
In this report, NH3, ClF and their constituent elements were investigated. Those molecules were optimized in Gaussian, then viewed and analysed in GaussView. The structural information including bond angle, bond distant, point group of those molecule were calculated. The optimal bond energy and the RMS gradient of those molecules were also calculated. The calculation method used was RB3LYP and the basis set used was 6-31G(d,p). The reaction energies of formation of NH3 and ClF were calculated from the bond energies. The vibration modes and molecular orbitals of those molecule were analysed in GaussView as well.
Ammonia NH3
Structure
| N-H Bond Distance | H-N-H Bond Angle | Point Group |
| 1.01798 A | 105.741o | C3v |
Ammonia |
The optimisation file is linked to here
Optimization Calculation Summery
| Molecule | Calculation Method | Basis Set | Final Energy (a.u.) | RMS Gradient (a.u.) |
| NH3 | RB3LYP | 6-31G(d,p) | -56.55776873 | 0.00000485 |
Data for the final set of forces and displacements
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986284D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
Vibration Modes

According to the 3N-6 rule: 3*4-6 = 6. There are 6 vibration modes for NH3 molecule. Mode 2 & 3 are degenerate. Mode 5 & 6 are degenerate. They have the same frequency, hence the same energy. Mode 1, 2, 3 are "bending" vibrations. Mode 4, 5, 6 are "bond stretch" vibrations. Mode 4 are highly symmetric because the symmetry of the molecule stays the same throughout the vibration. Mode 1 is known as the "umbrella" mode. 2 bands are expected to see in an experimental spectrum of gaseous ammonia. Because the intensities of Mode 4, 5, 6 are too small to be seen on the spectrum. This is due to those 3 vibrations have very small change in dipole moment.
Charge Distribution
| Hydrogen | Nitrogen |
| 0.375 | -1.125 |
N is expected to have a negative charge and H is expected to have a positive charge. Because nitrogen is more electronegative than hydrogen, it draws electrons towards itself. Therefore nitrogen has a negative charge.
Reaction Energies
Summary information of constituent element
| Bond Distance | Point Group | Calculation Method | Basis Set | Final Energy (a.u.) | RMS Gradient (a.u.) | Vibration Frequency (cm-1) | Charge On Atom |
| 1.10550 A | D∞h | RB3LYP | 6-31G(d.p) | -109.52412868 | 0.00000060 | 2457.33 | 0 |
Nitrogen is a homonuclear diatomic molecule, consists of two identical atoms which have the same electronegativity. Therefore the charge on each atom is 0. The intensity of vibration absorption is 0 since there is no change in dipole moment.
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.383741D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Nitrogen |
The optimisation file is linked to here
| Bond Distance | Point Group | Calculation Method | Basis Set | Final Energy (a.u.) | RMS Gradient (a.u.) | Vibration Frequency (cm-1) | Charge On Atom |
| 0.74279 A | D∞h | RB3LYP | 6-31G(d.p) | -1.17853936 | 0.00000017 | 4465.68 | 0 |
Hydrogen is a homonuclear diatomic molecule, consists of two identical atoms which have the same electronegativity. Therefore the charge on each atom is 0. The intensity of vibration absorption is 0 since there is no change in dipole moment.
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.167770D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Hydrogen |
The optimisation file is linked to here
Calculation for reaction energy
E(NH3)= -56.55776873 au
2*E(NH3)= -113.1155375 au
E(N2)= -109.52412868 au
E(H2)= -1.17853936 au
3*E(H2)= -3.53561808 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579074 au = -146.48 kJ
The energy released for converting 3 mol hydrogen and 1 mol nitrogen gas into 2 mol ammonia gas is 146.48 kJ. Since ΔE is negative, the ammonia product is more stable than the gaseous reactants.
Literature value = −46.2 kJ/mol[1]
- ↑ http://nshs-science.net/chemistry/common/pdf/R-standard_enthalpy_of_formation.pdf This is the E(NH3) reference.
2*-46.2 = -92.4 kJ
The calculated value is greater than the literature value.
Chlorine Monofluoride ClF
Structure
| Cl-F Bond Distance | Point Group |
| 1.66434 A | C∞v |
Chlorine Monofluoride |
The optimisation file is linked to here
Optimization Calculation Summery
| Molecule | Calculation Method | Basis Set | Final Energy (a.u.) | RMS Gradient (a.u.) |
| ClF | RB3LYP | 6-31G(d,p) | -559.94269578 | 0.00014211 |
Data for the final set of forces and displacements
Item Value Threshold Converged?
Maximum Force 0.000246 0.000450 YES
RMS Force 0.000246 0.000300 YES
Maximum Displacement 0.000433 0.001800 YES
RMS Displacement 0.000612 0.001200 YES
Predicted change in Energy=-9.606109D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.6643 -DE/DX = -0.0002 !
--------------------------------------------------------------------------------
Vibration Modes
According to the 3N-5 rule for linear molecules: 3*2-5 = 1. One vibration is expected for ClF.
Charge Distribution
| Chlorine | Fluorine |
| 0.309 | -0.309 |
F is expected to have a negative charge and Cl is expected to have a positive charge. Because fluorine is more electronegative than chlorine, it draws electrons towards itself. Therefore fluorine has a negative charge. The magnitude of the charge is the same, since ClF is neutral, the total charge is 0.
Reaction Energies
Summary information of constituent element
| Bond Distance | Point Group | Calculation Method | Basis Set | Final Energy (a.u.) | RMS Gradient (a.u.) | Vibration Frequency (cm-1) | Charge On Atom |
| 2.04174 A | D∞h | RB3LYP | 6-31G(d.p) | -920.34987886 | 0.00002511 | 520.32 | 0 |
Chlorine is a homonuclear diatomic molecule, consists of two identical atoms which have the same electronegativity. Therefore the charge on each atom is 0. The intensity of vibration absorption is 0 since there is no change in dipole moment.
Item Value Threshold Converged?
Maximum Force 0.000043 0.000450 YES
RMS Force 0.000043 0.000300 YES
Maximum Displacement 0.000121 0.001800 YES
RMS Displacement 0.000172 0.001200 YES
Predicted change in Energy=-4.911394D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.0417 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Chlorine |
The optimisation file is linked to here
| Bond Distance | Point Group | Calculation Method | Basis Set | Final Energy (a.u.) | RMS Gradient (a.u.) | Vibration Frequency (cm-1) | Charge On Atom |
| 1.40281 A | D∞h | RB3LYP | 6-31G(d.p) | -199.49825218 | 0.00007365 | 1065.09 | 0 |
Flurine is a homonuclear diatomic molecule, consists of two identical atoms which have the same electronegativity. Therefore the charge on each atom is 0. The intensity of vibration absorption is 0 since there is no change in dipole moment.
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000128 0.000300 YES
Maximum Displacement 0.000157 0.001800 YES
RMS Displacement 0.000221 0.001200 YES
Predicted change in Energy=-2.131104D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4028 -DE/DX = 0.0001 !
--------------------------------------------------------------------------------
Fluorine |
The optimisation file is linked to here
Calculation for reaction energy
E(ClF)= -559.94269578 au
E(Cl2)= -920.34987886 au
0.5*E(Cl2)= -460.1749394 au
E(F2)= -199.49825218 au
0.5*E(F2)= -99.74912609 au
ΔE=E(ClF)-[0.5*E(Cl2)+0.5*E(F2)]= -0.01863029 au = -48.91 kJ/mol
The energy released for converting 0.5 mol chlorine and 0.5 mol fluorine gas into 1 mol chlorine monofluoride is 48.91 kJ/mol. Since ΔE is negative, the product is more stable than the gaseous reactants.
Literature value = −56.5 kJ/mol[1]
- ↑ https://en.wikipedia.org/wiki/Chlorine_monofluoride This is the E(ClF) reference.
The calculated value is close to the literature value.
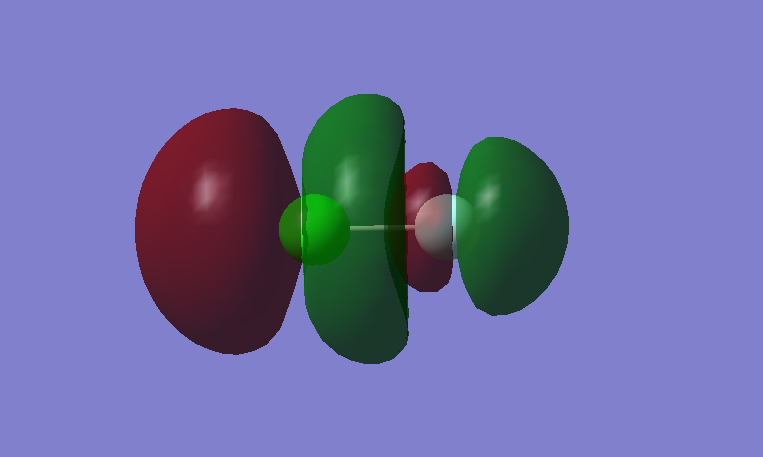
Molecular Orbitals
This is a bonding MO. The 2s orbital of fluorine and 3s orbital of chlorine contribute to this MO. This MO is occupied and it contributes to the bonding between the atoms. Its energy lies in the mid part of the energy levels.
This is an anti-bonding MO corresponding to the bonding MO above. The 2s orbital of fluorine and 3s orbital of chlorine contribute to this MO. This MO is occupied. It weakens the bonding between the atoms. Its energy lies in the mid part of the energy levels.
This is a bonding MO. The 2pz orbitals of fluorine and 3pz orbital of chlorine contribute to this MO. This MO is occupied and it contributes to the bonding of the atoms. The two lobes of the two p orbitals in-between the two atoms overlap and form a bond.
This is an anti-bonding MO corresponding to the bonding MO above. The 2pz orbitals of fluorine and 3pz orbital of chlorine contribute to this MO. This MO is unoccupied and it is the LUMO. If this MO is occupied, it would destabilize the bond between atoms.
This is a bonding MO. The 2px orbitals of fluorine and 3px orbital of chlorine contribute to this MO. This MO is occupied and it contributes to the bonding between the atoms.
This is an anti-bonding MO corresponding to the bonding MO above. The 2px orbitals of fluorine and 2px orbital of chlorine contribute to this MO. This MO is occupied and it is the HOMO. It weakens the bonding between the atoms.
There is one more bonding MO compared to anti-bonding MO, the bond order of this molecule is 1.