Rep:Mod:ZZ1617
NH3
'Item' Table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Information Table
| Item | Data |
|---|---|
| Molecule name | NH3 |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | 56.55776873 a.u. |
| Point Group | C3V |
Jmol dynamic image
test molecule |
The optimisation file is liked to here
Anmating vibrations
There are 6 modes expected according to the 3N-6 rule.
Modes with frequency 1693.95nm (#2 and #3 in the picture)are degenerate, Modes with frequency 3589.82nm (#5 and #6 in the picture) are degenerate.
Modes with frequencies 1089.54nm and 1693.95nm (#1, #2, and #3 in the picture) are "bending" vibrations, and modes with frequencies 3461.29nm and 3589.82nm (#4, #5, and #6 in the picture) are "bond stretch" vibrations.
Modes with frequencies 1089.54nm and 3461.29nm (#1 and #4 in the picture) are highly symmetric.
Mode with frequency 1089.54nm (#1 in the picture) is the "umbrella" mode.
4 bands (frequencies: 1089.54nm, 1693.95nm, 3461.29nm, and 3589.82nm) would be expected to see in an experimental spectrum of gaseous ammonia.
Atomic Charges
The charge on the N-atom is -1.125, and the charge on the H-atoms is 0.375.
I would expect a negative charge on the N-atom and positive charge on the H-atom because nitrogen is more electronegative than hydrogen.
N2
'Item' Table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Information Table
| Item | Data |
|---|---|
| Molecule name | N2 |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -109.52359111 a.u. |
| Point Group | D*H |
Jmol dynamic image
test molecule |
The optimisation file is liked to here
Anmating vibrations
| Mode # | Freq(nm) |
|---|---|
| 1 | 2457.33 |
H2
'Item' Table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Information Table
| Item | Data |
|---|---|
| Molecule name | H2 |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -1.17853936 a.u. |
| Point Group | D*H |
Jmol dynamic image
test molecule |
The optimisation file is liked to here
Anmating vibrations
| Mode # | Freq(nm) |
|---|---|
| 1 | 4465.68 |
Reaction Energies
Reaction: N2 + 3H2 -> 2NH3 E(NH3) = 56.55776873 a.u.
2*E(NH3) = 113.11553746 a.u.
E(N2) = 109.52359111 a.u.
E(H2) = 1.17853936 a.u.
3*E(H2) = 3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)] = 0.05632827 a.u. = 147.89 kJ/mol
Choice of small molecule: F2
'Item' Table
Item Value Threshold Converged? Maximum Force 0.000128 0.000450 YES RMS Force 0.000128 0.000300 YES Maximum Displacement 0.000156 0.001800 YES RMS Displacement 0.000221 0.001200 YES
Information Table
| Item | Data |
|---|---|
| Molecule name | F2 |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -199.49825218 a.u. |
| Point Group | D*H |
Jmol dynamic image
test molecule |
The optimisation file is liked to here
Anmating vibrations
| Mode # | Freq(nm) |
|---|---|
| 1 | 1065.09 |
Analysis: F2 molecule only has one mode with frequency 1065.09nm because it is a linear molecule (According to 3N-5 rule, 3*2-5=1 mode). The mode is a "bond stretch" vibration, and it is highly symmetric.
Charge Analysis
The charge for both atoms are 0 (i.e. they are not charged).
F2 molecule is not charged because it does not have any dipole moment, as expected.
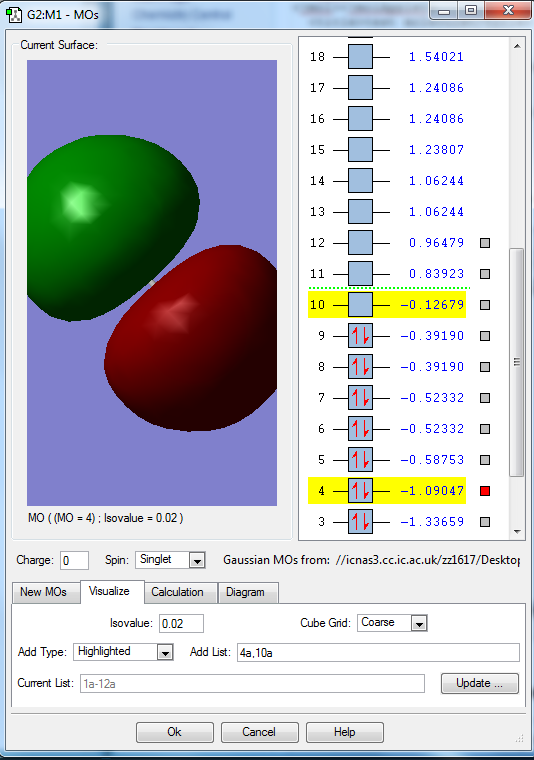
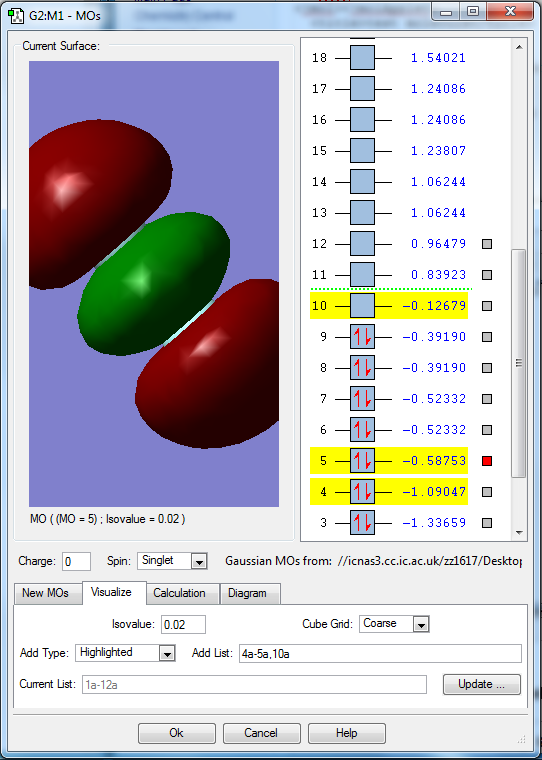
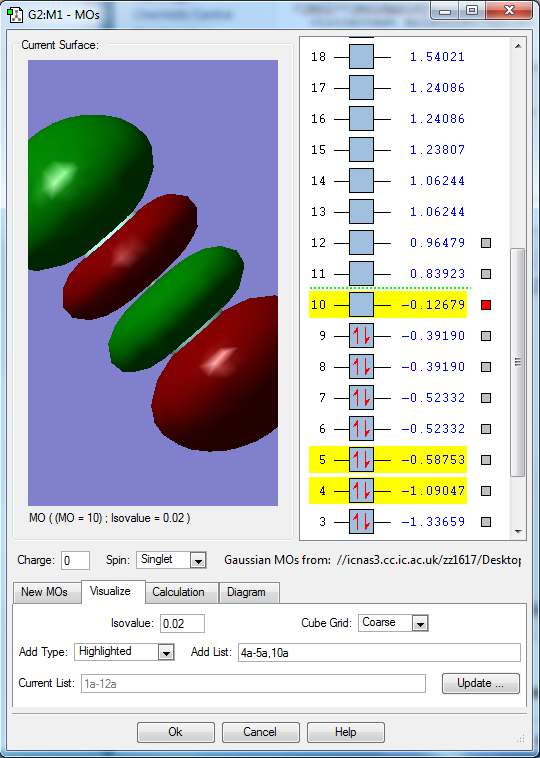
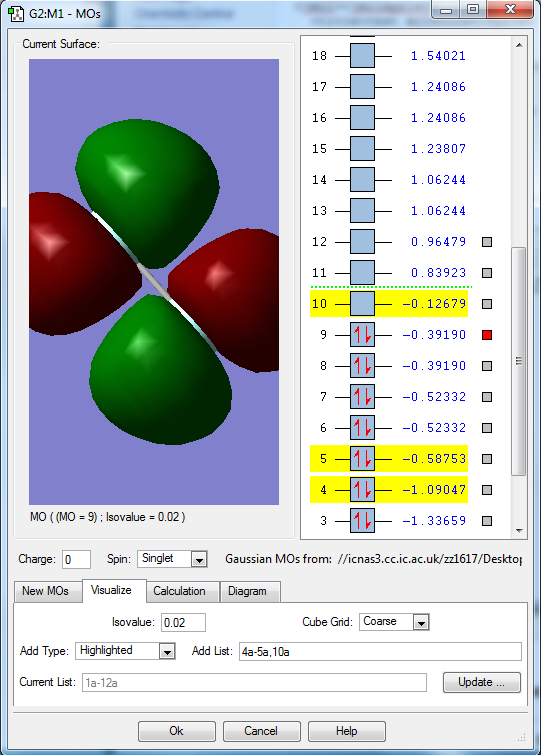
Molecular orbitals