Rep:Mod:ZQHMod2
In this module, Gaussian is used to predict the optimised structure of inorganic molecules. Also, the molecular orbital, the charge distribution and IR spectra of the optimised structure will be predicted. To obtain the optimised structure of the molecules, firstly the Schrodinger equation is solved for the electrons with an assumed position of nuclei. Born-Oppenheimer approximation is used, since electrons and nuclei are considered separately. This is called SCF part. Secondly the position of nuclei is calculated, which is OPT part. For each different position of nuclei, SCF is repeated until the lowest energy geometry is found. This is the optimised structure and the energy gradient is 0. In case of a maximum energy structure is obtained instead, the secondary derivate is calculated to ensure the result is a minimum point. In practise, the gradient is very small and close to 0.
Bonding (Ab initio and density functional molecular orbital)
BH3
Optimisation of BH3
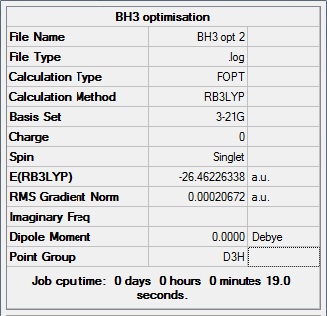
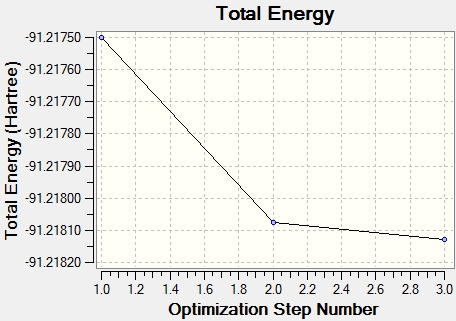
A BH3 molecule is created with Gaussian View5 with a trigonal planar structure, and 1.5A B-H bond length. The molecule is optimised with B3LYP method and 3-21G basic set. This setup will give out a result quickly with low accuracy. The BH3 molecule is a very simple one, for choosing this setup.
The information from .log file is shown.
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
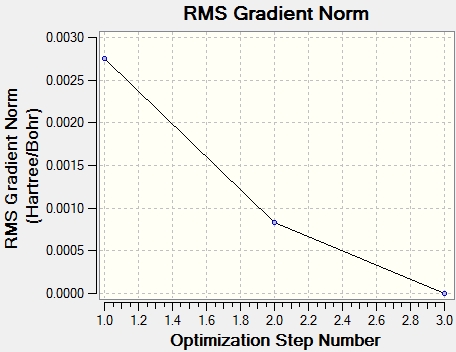
Item Value Threshold Converged? Maximum Force 0.000413 0.000450 YES RMS Force 0.000271 0.000300 YES Maximum Displacement 0.001610 0.001800 YES RMS Displacement 0.001054 0.001200 YES Predicted change in Energy=-1.071764D-06 Optimization completed. -- Stationary point found.
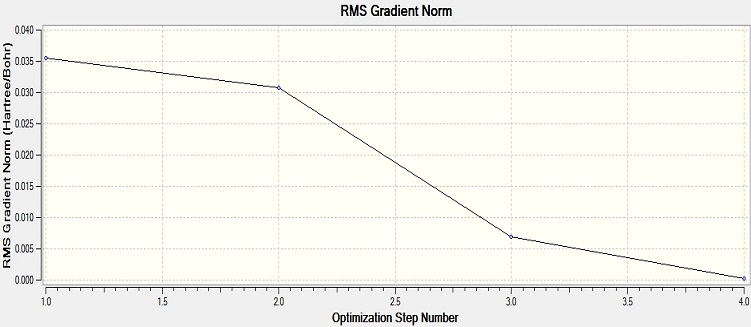
From the graph and results, it can be seen that the optimisation is successful, since the RMS gradient is very close to 0. From the molecule window of Gaussian View, the resultant B-H bond length is 1.19A and bong angle is 120°. And the dipole moment of the molecule is 0, due to its symmetry. It belongs to D3H group.
Frequency Analysis
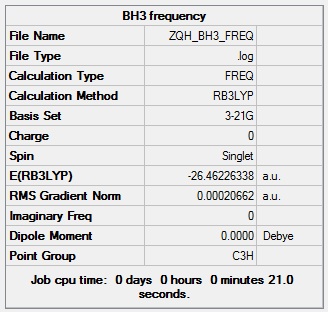
Frequency analysis of BH3 is used to predict its IR spectrum and to show the structure is at lowest energy. The spectrum will give negative values if the structure is not optimised well or the structure is a transition state with a maximum energy value. From the tabular results, the optimisation is good.
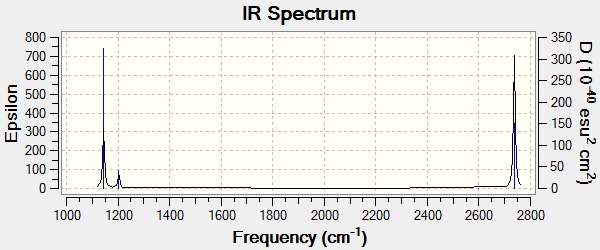
The frequency values are good, since the values of low frequencies are close to 0. There are six vibration frequencies for BH3, but only three peaks shown on the spectrum. The intensity of the peaks can explain one. The symmetric B-H stretch has 0 intensity, since the symmetry and the dipole moment of BH3 is 0. There are two pairs of values are degenerated, so only one peak will show. Therefore only three peaks are on the spectrum.
The point group shown on the summary is not correct since BH3 belongs to D3H group not C3H. The bond angle of H-B-H is 120 degree, which is same as the results from optimisation.
Low frequencies --- -66.7625 -66.3592 -66.3589 -0.0020 0.0031 0.2123 Low frequencies --- 1144.1483 1203.6413 1203.6424
| No. | Form of vibration | Frequency | Intensity | Description | .gif file |
| 1 | 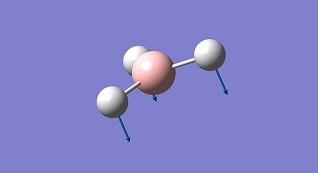
|
1144.15 | 98.9 | Deformation A2" | Media:BH3 vibration 1 zqh.ogg |
| 2 | 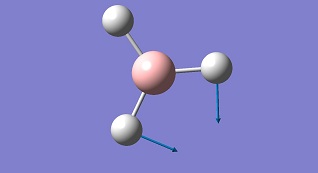
|
1203.64 | 12.3 | Symmetric B-H Bend E' | Media:BH3 vibration 2 zqh.ogg |
| 3 | 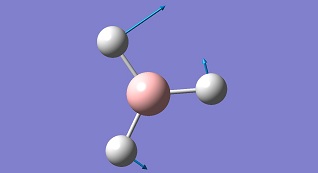
|
1203.64 | 12.3 | Asymmetric B-H Bend E' | Media:BH3 vibration 3 zqh.ogg |
| 4 | 
|
2598.42 | 0 | Symmetric B-H Stretch A1' | Media:BH3 vibration 4 zqh.ogg |
| 5 | 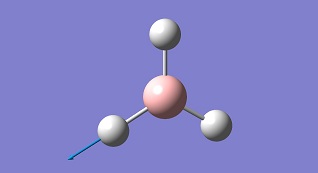
|
2737.44 | 103.7 | Asymmetric B-H Stretch E' | Media:BH3 vibration 5 zqh.ogg |
| 6 | 
|
2737.44 | 103.7 | Asymmetric B-H Stretch E' | Media:BH3 vibration 6 zqh.ogg |
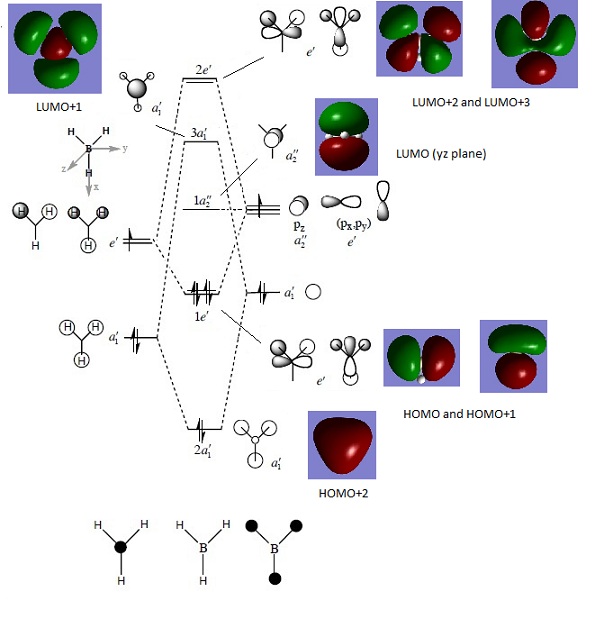
Molecular Orbital
The molecular orbital of the optimised structure is obtained by Energy analysis with parameter “pop=full” in the additional keyword section.
| Molecular orbital | Diagram | Energy |
| LUMO+3 | 
|
0.17954 |
| LUMO+2 | 
|
0.17954 |
| LUMO+1 | 
|
0.16635 |
| LUMO | 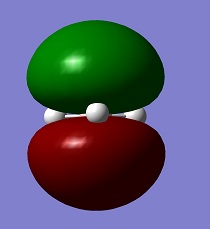
|
−0.06816 |
| HOMO | 
|
−0.35299 |
| HOMO-1 | 
|
−0.35299 |
| HOMO-2 | 
|
−0.51531 |
| HOMO-3 | 
|
−6.76609 |
The MO diagram of BH3 is drawn with the reference of the molecular orbital tutorial. The symmetry of the fragment orbitals are labelled and they are mixed with the orbitals with same symmetry and similar energy. The shape of the resultant MOs are same with the MOs calculated with Gaussian. But it is difficult to predict whether the 3a1' orbital has the higher energy than the degenerate 2e' orbitals or not from the MO diagram. The energy values calculated from Gaussian gives a reasonable prediction of the positions of unoccupied orbitals, and the occupied orbitals energy order is agreed with the prediction from the MO diagram. The energy of 1a1' orbital is too small, so it is not on the MO diagram.
NBO Analysis
The natural bond order of BH3 can be analysis by the same method as MO analysis, with additional keyword "pop=(full,nbo)". Gaussian View5 can use the density of different colours to show the charge on each atom.
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
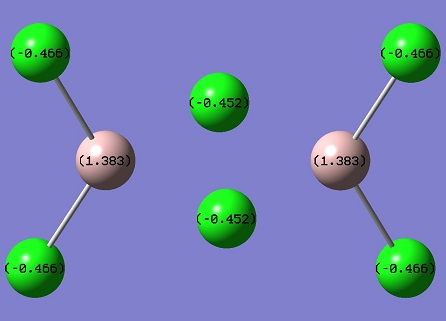
B 1 0.27816 1.99954 2.72230 0.00000 4.72184
H 2 -0.09272 0.00000 1.09256 0.00015 1.09272
H 3 -0.09272 0.00000 1.09256 0.00015 1.09272
H 4 -0.09272 0.00000 1.09256 0.00015 1.09272
=======================================================================
* Total * 0.00000 1.99954 6.00000 0.00046 8.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99854) BD ( 1) B 1 - H 2
( 45.36%) 0.6735* B 1 s( 33.33%)p 2.00( 66.67%)
0.0000 0.5774 0.0000 0.0000 0.0000
0.8165 0.0000 0.0000 0.0000
( 54.64%) 0.7392* H 2 s(100.00%)
1.0000 0.0001
2. (1.99854) BD ( 1) B 1 - H 3
( 45.36%) 0.6735* B 1 s( 33.33%)p 2.00( 66.67%)
0.0000 0.5774 0.0000 0.7071 0.0000
-0.4082 0.0000 0.0000 0.0000
( 54.64%) 0.7392* H 3 s(100.00%)
1.0000 0.0001
3. (1.99854) BD ( 1) B 1 - H 4
( 45.36%) 0.6735* B 1 s( 33.33%)p 2.00( 66.67%)
0.0000 0.5774 0.0000 -0.7071 0.0000
-0.4082 0.0000 0.0000 0.0000
( 54.64%) 0.7392* H 4 s(100.00%)
1.0000 0.0001
4. (1.99954) CR ( 1) B 1 s(100.00%)
1.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000
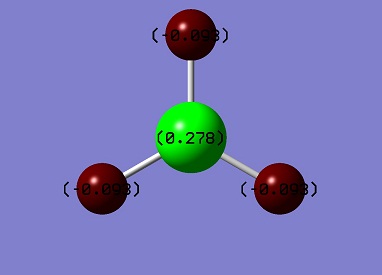
The graph shows the three H atoms have the same amount of charge, since the high symmetry of the molecule and the dipole moment of the molecule is 0. Boron is positively charged and Hydrogens are negatively charged. The 45% of charge density is on H and 55% of that on B. Also B-H bond has 33% s character and 67% p character, which indicates the sp2 hybridised bonds. It agrees the bond angle value = 120°.
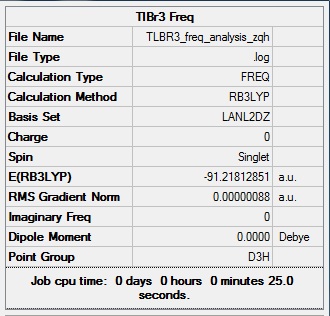
TlBr3
Optimisation of TlBr3
A TlBr3 molecule is created with Gaussian View5 with a trigonal planar structure. The molecule is optimised with B3LYP method and LANL2DZ basic set. This basis set uses psuedo potentials which is necessary for big atoms. Though LANL2DZ is another approximation, the setup will give a relative accurate result for large atoms like Tl.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Item Value Threshold Converged? Maximum Force 0.000002 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000022 0.001800 YES RMS Displacement 0.000014 0.001200 YES Predicted change in Energy=-6.083995D-11 Optimization completed. -- Stationary point found.
The RMS of the optimised structure is very close to 0, so the optimisation is successful. From the molecule window of Gaussian View, the resultant Tl-Br bond length is 2.56 A and bong angle is 120°. And the dipole moment of the molecule is 0, due to its symmetry. It belongs to D3H group. This molecule is also sp2 hybridised.
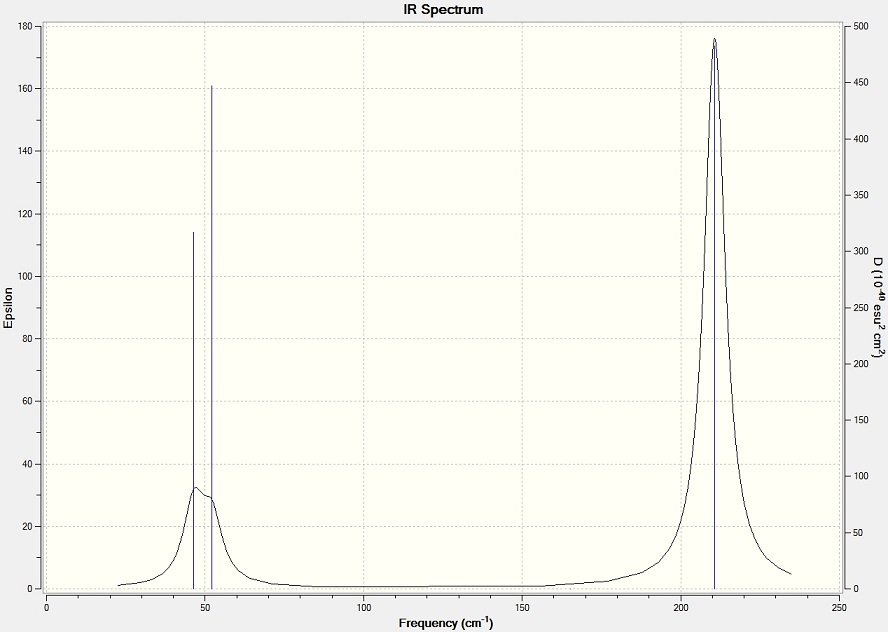
Frequency Analysis
The frequency analysis of TlBr3 shows the optimisation is successful, since no big negative value of absorption. There are six vibration frequencies, corresponding to six different modes of vibration. But there are only two main peaks. The intensity of the low frequencies are small, also two of them are degenerate. The completely symmetric stretch of Tl-Br bond gives 0 intensity. And finally the asymmetric stretch of Tl-Br bond shows two vibration modes with same energy level. So only two peaks are on the spectrum.
| No. | Form of vibration | Frequency | Intensity | Description | .gif file |
| 1 | 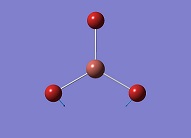
|
46.43 | 3.69 | Symmetric Tl-Br Bend E' | Media:TlBr3 freq 1 zqh.ogg |
| 2 | 
|
46.43 | 3.69 | Asymmetric Tl-Br Bend E' | Media:TlBr3 freq 2 zqh.ogg |
| 3 | 
|
52.14 | 5.85 | Deformation A2" | Media:TlBr3 freq 3 zqh.ogg |
| 4 | 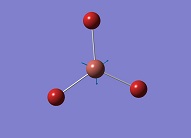
|
165.27 | 0 | Symmetric Tl-Br Stretch A1' | Media:TlBr3 freq 4 zqh.ogg |
| 5 | 
|
210.69 | 25.5 | Asymmetric Tl-Br Stretch E' | Media:TlBr3 freq 5 zqh.ogg |
| 6 | 
|
210.69 | 25.5 | Asymmetric Tl-Br Stretch E' | Media:TlBr3 freq 6 zqh.ogg |
The low frequencies shown blow are close to 0. This means the frequency analysis results are accurate.
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
Isomers of Mo(CO)4L2
The cis and trans forms of Mo(CO)4L2 are optimised with different basic set, LANL2MB and LANL2DZ. Then the vibration frequency analysis of the most stable structure is carried. The ligands attached on the central metal are PCl3. PPh3 is very common but it has more nuclei and electrons than PCl3. They have similar steric demand and PCl3 can show simpler bonding.
Optimisation of Mo(CO)4(PCl3)2
LANL2MB basic set
The molecules are optimised with B3LYP method and LANL2MB basic set. The addition keyword is filled with “opt=loose” to obtain a loose optimisation.
cis Mo(CO)4(PCl3)2
Item Value Threshold Converged? Maximum Force 0.000080 0.002500 YES RMS Force 0.000021 0.001667 YES Maximum Displacement 0.008443 0.010000 YES RMS Displacement 0.002151 0.006667 YES Predicted change in Energy=-9.900242D-08 Optimization completed. -- Stationary point found.
trans Mo(CO)4(PCl3)2
Item Value Threshold Converged? Maximum Force 0.000164 0.002500 YES RMS Force 0.000054 0.001667 YES Maximum Displacement 0.004456 0.010000 YES RMS Displacement 0.001407 0.006667 YES Predicted change in Energy=-7.803340D-07 Optimization completed. -- Stationary point found.
LanL2DZ Basic Set
The molecules are optimised with B3LYP method and LanL2DZ basic set. The Dihedral Angles of the molecules are adjusted before the optimisation. The trans isomer is adjusted to the form with eclipsed PCl3 groups. The Dihedral Angle between two Cl atom on different P is 180°. The Dihedral Angle between a Cl atom on one P and a C atom of the cis isomer is 180°and Dihedral Angle between a Cl atom on the other P and the same C is 0°. The addition keyword is filled with “int=unltrafine scf=conver=9” to increase the electronic convergence.
cis Mo(CO)4(PCl3)2
Item Value Threshold Converged? Maximum Force 0.000040 0.000450 YES RMS Force 0.000017 0.000300 YES Maximum Displacement 0.001685 0.001800 YES RMS Displacement 0.000556 0.001200 YES Predicted change in Energy=-8.926135D-08 Optimization completed. -- Stationary point found.
trans Mo(CO)4(PCl3)2
Item Value Threshold Converged? Maximum Force 0.000097 0.000450 YES RMS Force 0.000023 0.000300 YES Maximum Displacement 0.001668 0.001800 YES RMS Displacement 0.000568 0.001200 YES Predicted change in Energy=-1.073575D-07 Optimization completed. -- Stationary point found.
LanL2DZ Basis Set with extra basic set
The .log file is changed. The d orbital functions of P is added manually.
cis Mo(CO)4(PCl3)2
Item Value Threshold Converged? Maximum Force 0.000081 0.000450 YES RMS Force 0.000023 0.000300 YES Maximum Displacement 0.001121 0.001800 YES RMS Displacement 0.000401 0.001200 YES Predicted change in Energy=-7.863202D-08 Optimization completed. -- Stationary point found.
trans Mo(CO)4(PCl3)2
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000074 0.001800 YES RMS Displacement 0.000024 0.001200 YES Predicted change in Energy=-2.571683D-10 Optimization completed. -- Stationary point found.
| Bond type | Bond length (A) | Bond length (A) |
| cis Mo(CO)4(PCl3)2 | trans Mo(CO)4(PCl3)2 | |
| CO | 1.17-1.18 | 1.17 |
| MoC | 2.02-2.05 | 2.06 |
| MoP | 2.48 | 2.42 |
| PCl | 2.12 | 2.12 |
The CO and PCl bond length of two isomers are same. The cis isomer has a longer MoP bond length than that of trans isomer, since trans isomer has a lower steric effect. The two PCl3 group will suffer smaller repel force from each other. Due to this, the trans isomer has a longer MoC bond length. In cis isomer, the MoC bond for the carbonyl group, which the Mo-C-P bond angle for the same C and different P are both 90 degree, is longer than that of the other two MoC bond.
The trans isomer is thermodynamically more stable with a energy. The 2nd year synthesis lab also confirms this. At a certain temperature, the cis isomer would like to turn into trans isomer. Without taking account the d orbitals of P using extra basic set, the result of optimisation will say that the cis isomer is more stable than the trans one.
Frequency analysis
The structure with the lowest energy of each isomer is used for frequency analysis.
cis Mo(CO)4(PCl3)2
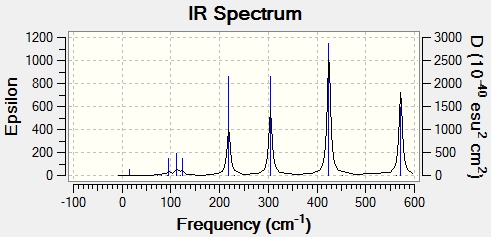
| No. | Form of vibration | Frequency | Intensity | Description | .gif file |
| 1 | 
|
1937.72 | 1574 | Asymmetric C=O stretch B2' | Media:cis Mo(CO)4(PCl3)2 freq 1 zqh.ogg |
| 2 | 
|
1940.36 | 840.5 | Asymmetric C=O stretch B2 | Media:cis Mo(CO)4(PCl3)2 freq 2 zqh.ogg |
| 3 | 
|
1952.01 | 606.8 | Asymmetric C=O stretch A1' | Media:cis Mo(CO)4(PCl3)2 freq 3 zqh.ogg |
| 4 | 
|
2018.94 | 558.1 | Symmetric C=O stretch A1' | Media:cis Mo(CO)4(PCl3)2 freq 4 zqh.ogg |
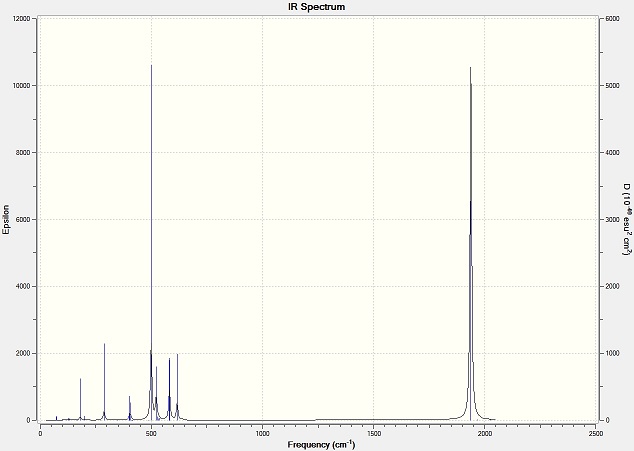
For CO groups of cis isomer, four vibration modes generate four peaks on the graph. The molecule is at lower symmetry, so all the stretches of CO group have high intensity and shown on the spectrum.
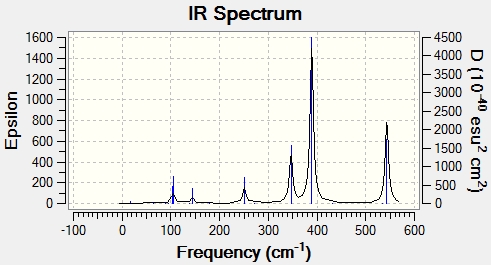
Media:trans Mo(CO)4(PCl3)2 freq 1 zqh.ogg trans Mo(CO)4(PCl3)2
| No. | Form of vibration | Frequency | Intensity | Description | .gif file |
| 1 | 
|
1937.85 | 1590 | Asymmetric C=O stretch Eu | Media:trans Mo(CO)4(PCl3)2 freq 1 zqh.ogg |
| 2 | 
|
1938.52 | 1587 | Asymmetric C=O stretch Eu | Media:trans Mo(CO)4(PCl3)2 freq 2 zqh.ogg |
| 3 | 
|
1966.60 | 3.45 | Asymmetric C=O stretch B1g | Media:trans Mo(CO)4(PCl3)2 freq 3 zqh.ogg |
| 4 | 
|
2024.26 | 5.43 | Symmetric C=O stretch A1g | Media:trans Mo(CO)4(PCl3)2 freq 4 zqh.ogg |
For CO groups of trans isomer, four vibration modes generate one peak on the graph. The high symmetry of the molecule makes the symmetric stretches of CO to have a low intensity. And the asymmetric stretches of CO are degenerate, so only one peak is shown on the spectrum.
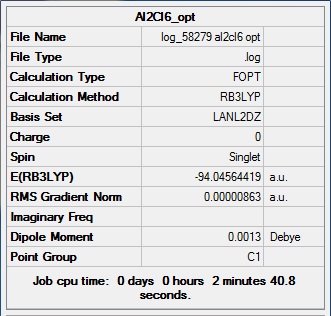
Mini Project
Al2Cl6 shows a dimmer form. The two Cl atoms are the bridge atoms between Al. If two Cl atoms are substituted with two Br atoms, there are three different resultant isomers: the two Br atoms are the bridge atoms, the two Br atoms are on the same side and the two Br atoms are on the different side of the molecule. To find the most stable and thermodynamically favoured resultant molecule, the four atoms will be optimised first. The molecular orbitals, natural bond order and vibrational frequency will be analysis later.
Optimisation
The four molecules are drawn with Gaussian View 5 and optimised with B3LYP method and LanL2DZ basic set to obtain a loose optimisation. Then the additional words “int=ultrafine scf=cover=9” are added to optimise the molecules from the pre-optimised structure. The results below show the final optimisation step.
Al2Cl6
Item Value Threshold Converged? Maximum Force 0.000015 0.000450 YES RMS Force 0.000007 0.000300 YES Maximum Displacement 0.000987 0.001800 YES RMS Displacement 0.000276 0.001200 YES Predicted change in Energy=-1.506461D-08 Optimization completed. -- Stationary point found.
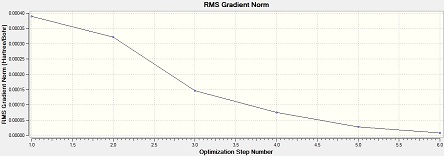
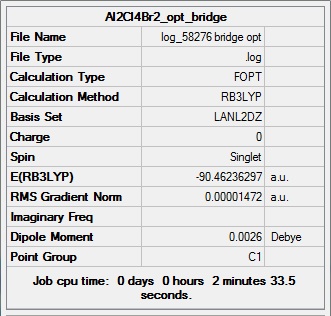
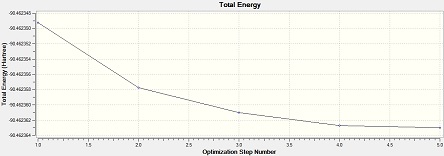
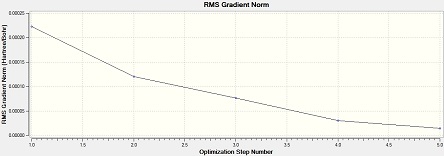
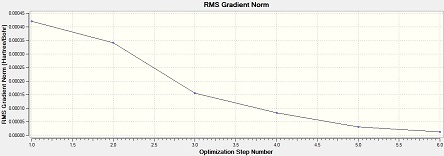
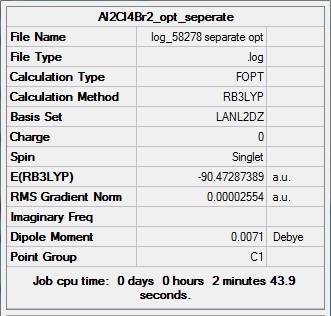
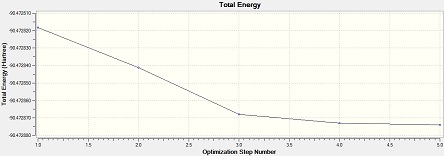
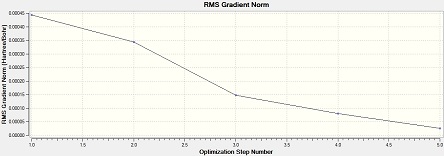
Al2Cl4Br2 (bridge Br)
Item Value Threshold Converged? Maximum Force 0.000029 0.000450 YES RMS Force 0.000013 0.000300 YES Maximum Displacement 0.000793 0.001800 YES RMS Displacement 0.000338 0.001200 YES Predicted change in Energy=-1.563712D-08 Optimization completed. -- Stationary point found.
AlCl4Br2 (same end Br)
Item Value Threshold Converged? Maximum Force 0.000034 0.000450 YES RMS Force 0.000013 0.000300 YES Maximum Displacement 0.001486 0.001800 YES RMS Displacement 0.000381 0.001200 YES Predicted change in Energy=-3.875085D-08 Optimization completed. -- Stationary point found.
Al2Cl4Br2 (different ends trans Br)
Item Value Threshold Converged? Maximum Force 0.000043 0.000450 YES RMS Force 0.000019 0.000300 YES Maximum Displacement 0.001535 0.001800 YES RMS Displacement 0.000447 0.001200 YES Predicted change in Energy=-7.970282D-08 Optimization completed. -- Stationary point found.
Frequency analysis
From the low frequencies values, the optimisation is successful.
Al2Cl4Br2 (bridge Br)
Low frequencies --- -1.7473 -1.5905 -0.9962 0.0000 0.0001 0.0001 Low frequencies --- 15.8976 52.6261 72.9895
AlCl4Br2 (same end Br)
Low frequencies --- 0.0000 0.0000 0.0000 0.8585 1.1383 2.4394 Low frequencies --- 18.2658 42.1513 64.3173
Al2Cl4Br2 (different ends trans Br)
Low frequencies --- -3.0440 -1.4037 -1.2127 0.0000 0.0000 0.0000 Low frequencies --- 17.9006 40.6517 64.4989
Al2Cl6
Low frequencies --- -0.9559 -0.2690 -0.0003 0.0000 0.0003 1.6891 Low frequencies --- 21.3598 51.4303 84.9699
The IR spectra of the substituted molecules are different from the original molecule. Only the vibrations with high intensity are analysed. Al2Cl4Br2 have low frequencies than Al2Cl6, especially the vibration modes involves the movement of Br atoms. The reason for this is the bond length involves Br atom is longer, so the vibration frequency is smaller, indicating a weaker bond. If the vibration modes only contain Al and Cl atoms, the frequency values are similar as Al2Cl6. These can be seen from the table below. The Al2Cl4Br2 molecules with two Br atoms at same end are quite different. It looses the symmetry element “i”, the inversion centre, so it shows more peaks due to the asymmetric vibrations. The addition peaks can be paired with the originally existing peaks, which have the similar vibration frequencies as Al2Cl6.
NBO analysis
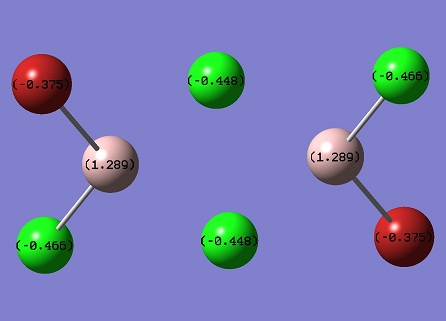
Al atoms carry positive charge, while Cl and Br carry negative charge. Cl always carries more charge than Br, since its electronegativity is higher than that of Br. If Al is bonding with Br, the charge carried will be smaller. Bridging Cl and Br atoms will carry less charge than ending atoms, since coordinate bond makes them loose electron density.
Al2Cl4Br2 (bridge Br)
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.96604) BD ( 1)Al 1 -Cl 2
( 17.82%) 0.4221*Al 1 s( 31.08%)p 2.22( 68.92%)
0.5575 0.0080 0.4331 0.0379 0.7038
0.0696 0.0001 0.0000
( 82.18%) 0.9066*Cl 2 s( 22.24%)p 3.50( 77.76%)
0.4716 0.0004 -0.4340 -0.0048 -0.7676
-0.0072 -0.0002 0.0000
2. (1.96604) BD ( 1)Al 1 -Cl 3
( 17.82%) 0.4221*Al 1 s( 31.08%)p 2.22( 68.92%)
0.5575 0.0080 0.4334 0.0380 -0.7036
-0.0696 0.0001 0.0000
( 82.18%) 0.9066*Cl 3 s( 22.24%)p 3.50( 77.76%)
0.4716 0.0004 -0.4344 -0.0048 0.7674
0.0072 -0.0002 0.0000
3. (1.95588) BD ( 1)Al 1 -Br 7
( 14.35%) 0.3789*Al 1 s( 18.88%)p 4.30( 81.12%)
0.4343 -0.0124 -0.5576 -0.0132 -0.0001
0.0000 0.7057 0.0463
( 85.65%) 0.9255*Br 7 s( 17.27%)p 4.79( 82.73%)
0.4156 0.0001 0.7070 -0.0001 0.0000
0.0000 -0.5722 -0.0012
4. (1.95588) BD ( 1)Al 1 -Br 8
( 14.37%) 0.3790*Al 1 s( 18.90%)p 4.29( 81.10%)
0.4346 -0.0123 -0.5576 -0.0133 -0.0001
0.0000 -0.7055 -0.0464
( 85.63%) 0.9254*Br 8 s( 17.27%)p 4.79( 82.73%)
0.4156 0.0001 0.7072 -0.0001 0.0000
0.0000 0.5720 0.0012
5. (1.96603) BD ( 1)Al 4 -Cl 5
( 17.81%) 0.4221*Al 4 s( 31.08%)p 2.22( 68.92%)
0.5575 0.0080 -0.4330 -0.0379 0.7038
0.0696 -0.0001 0.0000
( 82.19%) 0.9066*Cl 5 s( 22.24%)p 3.50( 77.76%)
0.4716 0.0004 0.4338 0.0048 -0.7677
-0.0072 0.0003 0.0000
6. (1.96604) BD ( 1)Al 4 -Cl 6
( 17.82%) 0.4221*Al 4 s( 31.09%)p 2.22( 68.91%)
0.5575 0.0080 -0.4335 -0.0380 -0.7035
-0.0696 -0.0001 0.0000
( 82.18%) 0.9066*Cl 6 s( 22.24%)p 3.50( 77.76%)
0.4716 0.0004 0.4346 0.0048 0.7672
0.0072 0.0004 0.0000
7. (1.95588) BD ( 1)Al 4 -Br 7
( 14.37%) 0.3790*Al 4 s( 18.90%)p 4.29( 81.10%)
-0.4346 0.0123 -0.5576 -0.0133 0.0002
0.0000 -0.7055 -0.0464
( 85.63%) 0.9254*Br 7 s( 17.27%)p 4.79( 82.73%)
-0.4156 -0.0001 0.7072 -0.0001 0.0000
0.0000 0.5720 0.0012
8. (1.95588) BD ( 1)Al 4 -Br 8
( 14.35%) 0.3789*Al 4 s( 18.88%)p 4.30( 81.12%)
-0.4343 0.0124 -0.5576 -0.0132 0.0002
0.0000 0.7057 0.0463
( 85.65%) 0.9255*Br 8 s( 17.27%)p 4.79( 82.73%)
-0.4155 -0.0001 0.7071 -0.0001 0.0000
0.0000 -0.5722 -0.0012
AlCl4Br2 (same end Br)
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.96255) BD ( 1)Al 1 -Cl 2
( 11.85%) 0.3442*Al 1 s( 19.55%)p 4.12( 80.45%)
0.4419 -0.0143 -0.5553 -0.0158 -0.0009
-0.0003 0.7030 0.0424
( 88.15%) 0.9389*Cl 2 s( 21.10%)p 3.74( 78.90%)
0.4593 0.0026 0.7147 -0.0001 0.0038
0.0000 -0.5275 0.0009
2. (1.96230) BD ( 1)Al 1 -Cl 3
( 11.45%) 0.3384*Al 1 s( 18.86%)p 4.30( 81.14%)
0.4339 -0.0163 -0.5543 -0.0125 0.0005
-0.0002 -0.7088 -0.0406
( 88.55%) 0.9410*Cl 3 s( 21.02%)p 3.76( 78.98%)
0.4585 0.0026 0.7094 -0.0005 0.0041
0.0000 0.5353 -0.0016
3. (1.96635) BD ( 1)Al 1 -Cl 4
( 17.80%) 0.4220*Al 1 s( 30.75%)p 2.25( 69.25%)
0.5544 0.0105 0.4367 0.0401 -0.7035
-0.0730 -0.0034 -0.0008
( 82.20%) 0.9066*Cl 4 s( 21.57%)p 3.64( 78.43%)
0.4644 0.0010 -0.4271 -0.0047 0.7757
0.0072 0.0128 0.0001
4. (1.96628) BD ( 1)Al 1 -Cl 5
( 17.82%) 0.4222*Al 1 s( 30.81%)p 2.25( 69.19%)
-0.5550 -0.0103 -0.4364 -0.0406 -0.7032
-0.0728 0.0020 0.0006
( 82.18%) 0.9065*Cl 5 s( 21.53%)p 3.64( 78.47%)
-0.4640 -0.0009 0.4322 0.0047 0.7731
0.0072 -0.0099 0.0000
5. (1.96336) BD ( 1)Cl 2 -Al 6
( 88.67%) 0.9417*Cl 2 s( 22.79%)p 3.39( 77.21%)
-0.4774 -0.0020 0.6994 -0.0006 -0.0062
0.0000 0.5319 -0.0023
( 11.33%) 0.3366*Al 6 s( 18.04%)p 4.54( 81.96%)
-0.4242 0.0215 -0.5597 0.0004 0.0116
0.0006 -0.7112 -0.0206
6. (1.96357) BD ( 1)Cl 3 -Al 6
( 88.28%) 0.9396*Cl 3 s( 23.12%)p 3.33( 76.88%)
-0.4808 -0.0020 0.7046 -0.0002 -0.0057
0.0000 -0.5219 0.0016
( 11.72%) 0.3423*Al 6 s( 18.83%)p 4.31( 81.17%)
-0.4336 0.0188 -0.5637 -0.0036 0.0120
0.0007 0.7023 0.0230
7. (1.95796) BD ( 1)Al 6 -Br 7
( 21.97%) 0.4687*Al 6 s( 31.45%)p 2.18( 68.55%)
-0.5607 -0.0106 0.4126 0.0354 -0.7128
-0.0768 -0.0023 -0.0007
( 78.03%) 0.8833*Br 7 s( 18.09%)p 4.53( 81.91%)
-0.4253 -0.0004 -0.4106 -0.0085 0.8063
0.0133 0.0107 0.0001
8. (1.95826) BD ( 1)Al 6 -Br 8
( 22.00%) 0.4691*Al 6 s( 31.62%)p 2.16( 68.38%)
0.5622 0.0118 -0.4434 -0.0386 -0.6928
-0.0753 0.0026 0.0008
( 78.00%) 0.8831*Br 8 s( 18.11%)p 4.52( 81.89%)
0.4255 0.0004 0.4555 0.0092 0.7817
0.0128 -0.0119 -0.0001
Al2Cl4Br2 (different ends trans Br)
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.96286) BD ( 1)Al 1 -Cl 2
( 11.58%) 0.3403*Al 1 s( 18.82%)p 4.31( 81.18%)
0.4335 -0.0179 0.5193 0.0076 0.2053
0.0019 -0.7063 -0.0314
( 88.42%) 0.9403*Cl 2 s( 22.00%)p 3.55( 78.00%)
0.4690 0.0023 -0.6598 0.0003 -0.2547
0.0000 0.5290 -0.0016
2. (1.96285) BD ( 1)Al 1 -Cl 3
( 11.57%) 0.3402*Al 1 s( 18.80%)p 4.32( 81.20%)
-0.4333 0.0180 -0.5192 -0.0075 -0.2054
-0.0019 -0.7065 -0.0314
( 88.43%) 0.9404*Cl 3 s( 21.99%)p 3.55( 78.01%)
-0.4689 -0.0023 0.6596 -0.0003 0.2549
0.0000 0.5292 -0.0016
3. (1.96642) BD ( 1)Al 1 -Cl 4
( 17.76%) 0.4214*Al 1 s( 30.98%)p 2.23( 69.02%)
0.5565 0.0111 -0.1413 -0.0054 -0.8150
-0.0771 0.0001 0.0000
( 82.24%) 0.9069*Cl 4 s( 22.37%)p 3.47( 77.63%)
0.4730 0.0009 0.0994 0.0014 0.8754
0.0083 -0.0003 0.0000
4. (1.95774) BD ( 1)Al 1 -Br 8
( 21.97%) 0.4687*Al 1 s( 31.35%)p 2.19( 68.65%)
0.5598 0.0108 -0.6606 -0.0694 0.4920
0.0571 0.0000 0.0000
( 78.03%) 0.8834*Br 8 s( 17.46%)p 4.73( 82.54%)
0.4179 0.0003 0.7040 0.0134 -0.5740
-0.0087 -0.0003 0.0000
5. (1.96287) BD ( 1)Cl 2 -Al 5
( 88.42%) 0.9403*Cl 2 s( 22.01%)p 3.54( 77.99%)
0.4691 0.0023 0.6594 -0.0003 0.2550
0.0000 0.5293 -0.0016
( 11.58%) 0.3402*Al 5 s( 18.81%)p 4.32( 81.19%)
0.4333 -0.0179 -0.5193 -0.0075 -0.2052
-0.0019 -0.7065 -0.0314
6. (1.96288) BD ( 1)Cl 3 -Al 5
( 88.41%) 0.9403*Cl 3 s( 22.01%)p 3.54( 77.99%)
0.4691 0.0023 0.6595 -0.0003 0.2549
0.0000 -0.5291 0.0016
( 11.59%) 0.3404*Al 5 s( 18.83%)p 4.31( 81.17%)
0.4335 -0.0179 -0.5194 -0.0076 -0.2050
-0.0019 0.7064 0.0314
7. (1.96643) BD ( 1)Al 5 -Cl 6
( 17.76%) 0.4214*Al 5 s( 30.98%)p 2.23( 69.02%)
-0.5565 -0.0111 -0.1419 -0.0055 -0.8149
-0.0771 0.0002 0.0000
( 82.24%) 0.9069*Cl 6 s( 22.37%)p 3.47( 77.63%)
-0.4730 -0.0009 0.1004 0.0014 0.8753
0.0083 -0.0003 0.0000
8. (1.95775) BD ( 1)Al 5 -Br 7
( 21.97%) 0.4687*Al 5 s( 31.34%)p 2.19( 68.66%)
-0.5597 -0.0107 -0.6604 -0.0693 0.4924
0.0571 0.0000 0.0000
( 78.03%) 0.8834*Br 7 s( 17.46%)p 4.73( 82.54%)
-0.4179 -0.0003 0.7035 0.0133 -0.5747
-0.0087 -0.0001 0.0000
Al2Cl6
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.96291) BD ( 1)Al 1 -Cl 2
( 11.49%) 0.3390*Al 1 s( 18.85%)p 4.30( 81.15%)
-0.4339 0.0156 -0.5580 -0.0128 -0.0001
0.0000 -0.7060 -0.0394
( 88.51%) 0.9408*Cl 2 s( 21.22%)p 3.71( 78.78%)
-0.4607 -0.0026 0.7071 -0.0005 0.0000
0.0000 0.5364 -0.0017
2. (1.96291) BD ( 1)Al 1 -Cl 3
( 11.49%) 0.3389*Al 1 s( 18.84%)p 4.31( 81.16%)
-0.4338 0.0156 -0.5580 -0.0128 -0.0001
0.0000 0.7060 0.0394
( 88.51%) 0.9408*Cl 3 s( 21.22%)p 3.71( 78.78%)
-0.4607 -0.0026 0.7070 -0.0005 -0.0001
0.0000 -0.5365 0.0017
3. (1.96604) BD ( 1)Al 1 -Cl 4
( 17.90%) 0.4230*Al 1 s( 31.14%)p 2.21( 68.86%)
-0.5579 -0.0106 0.4327 0.0391 -0.7032
-0.0737 0.0000 0.0000
( 82.10%) 0.9061*Cl 4 s( 21.27%)p 3.70( 78.73%)
-0.4611 -0.0009 -0.4275 -0.0047 0.7775
0.0073 -0.0001 0.0000
4. (1.96604) BD ( 1)Al 1 -Cl 5
( 17.89%) 0.4230*Al 1 s( 31.13%)p 2.21( 68.87%)
0.5579 0.0106 -0.4325 -0.0391 -0.7033
-0.0737 0.0000 0.0000
( 82.11%) 0.9061*Cl 5 s( 21.27%)p 3.70( 78.73%)
0.4612 0.0009 0.4271 0.0047 0.7777
0.0073 0.0001 0.0000
5. (1.96290) BD ( 1)Cl 2 -Al 6
( 88.51%) 0.9408*Cl 2 s( 21.22%)p 3.71( 78.78%)
0.4606 0.0026 0.7071 -0.0005 -0.0001
0.0000 -0.5365 0.0017
( 11.49%) 0.3389*Al 6 s( 18.84%)p 4.31( 81.16%)
0.4338 -0.0156 -0.5580 -0.0128 -0.0002
0.0000 0.7061 0.0394
6. (1.96290) BD ( 1)Cl 3 -Al 6
( 88.51%) 0.9408*Cl 3 s( 21.22%)p 3.71( 78.78%)
0.4607 0.0026 0.7072 -0.0005 -0.0001
0.0000 0.5364 -0.0017
( 11.49%) 0.3390*Al 6 s( 18.85%)p 4.30( 81.15%)
0.4339 -0.0156 -0.5580 -0.0128 -0.0002
0.0000 -0.7060 -0.0394
7. (1.96604) BD ( 1)Al 6 -Cl 7
( 17.89%) 0.4230*Al 6 s( 31.13%)p 2.21( 68.87%)
-0.5579 -0.0106 -0.4323 -0.0391 -0.7034
-0.0737 0.0000 0.0000
( 82.11%) 0.9061*Cl 7 s( 21.27%)p 3.70( 78.73%)
-0.4611 -0.0009 0.4270 0.0047 0.7778
0.0073 0.0001 0.0000
8. (1.96604) BD ( 1)Al 6 -Cl 8
( 17.90%) 0.4230*Al 6 s( 31.14%)p 2.21( 68.86%)
0.5579 0.0106 0.4328 0.0391 -0.7031
-0.0737 0.0000 0.0000
( 82.10%) 0.9061*Cl 8 s( 21.27%)p 3.70( 78.73%)
0.4611 0.0009 -0.4279 -0.0047 0.7773
0.0073 -0.0002 0.0000
In Al2Cl6 and Al2Cl4Br2, the bridging bonds have more p character than ending bonds. It indicates that the centre Al atoms are electron deficient, the bonding electrons from Cl or Br atoms are p lone pair electrons. So the p character increases. The Al atom is nearly sp2 hybridised. The ending bonds shows 30% s and 70% p roughly for all four molecules.
Conclusion
In the mini project, the structures of the molecules are optimised successfully, the IR spectra are predicted and charge distribution is illustrated. The bond type of the molecules calculated by Gaussian shows the donation of electrons to the Al atoms in the bridging bonds.
Conclusion
The calculation and prediction made by computational techniques are useful and accurate to analysis inorganic molecules. The optimisation can give out a correct geometry of the molecule. The frequency analysis can predict the shape of IR spectra. Also the NBO analysis and MO calculated by Gaussian can give a good view of the bonding in the molecules.
Reference
1 http://www.huntresearchgroup.org.uk/teaching/teaching_comp_lab_year3/Tut_MO_diagram_BH3.pdf
2 http://hdl.handle.net/10042/to-13605
3 http://hdl.handle.net/10042/to-13607
4 http://hdl.handle.net/10042/to-13606
5 http://hdl.handle.net/10042/to-13608
6 http://hdl.handle.net/10042/to-13609
7 http://hdl.handle.net/10042/to-13610
8 http://hdl.handle.net/10042/to-13369
9 http://hdl.handle.net/10042/to-13604
10 M. Y. Darensbourg and D. J. Darensbourg, J. Chem. Ed. 1970, 47, 33
11 D. J. Darensbourg and R. L. Kemp, Inorg. Chem. 1978, 17, 2680
12 http://hdl.handle.net/10042/to-13614
13 http://hdl.handle.net/10042/to-13613
14 http://hdl.handle.net/10042/to-13612
15 http://hdl.handle.net/10042/to-13611
16 http://hdl.handle.net/10042/to-13618
17 http://hdl.handle.net/10042/to-13617