Rep:Mod:ZL6217
Appearance
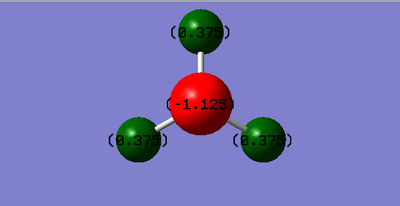
NH3 molecules
- molecule name: ammonia
- calculation method:RB3LYP
- basis set:6-31G(d,p)
- final energy E(RB3LYP): -56.55776873 a.u.
- the point group: C3V
- RMS gradient:0.00000323 a.u.
- optimised N-H bond distance: 1.01798
- optimised H-N-H bond angle: 105.745
test molecule |
The optimisation file is liked to here
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000014 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.141655D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7446 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7446 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7446 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8637 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
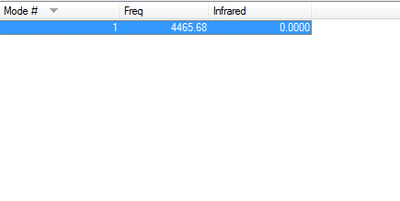
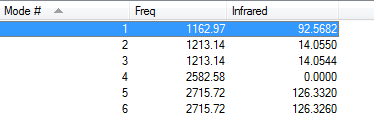
- Vibration:
- how many modes do you expect from the 3N-6 rule?
- Six
- which modes are degenerate (i.e. have the same energy)?
- Mode 2 and 3 / mode 5 and 6 are degenerate
- which modes are "bending" vibrations and which are "bond stretch" vibrations?
- mode 2 and 3
- which mode is highly symmetric?
- mode 1,3 and 4
- one mode is known as the "umbrella" mode, which one is this?
- mode 4
- how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
- 4
- Charge distribution:
- Charge on N-atom: -1.125
- Charge on H-atom: 0.375
- Expectation value: Negative charge on N-atom and positive charge on H-atoms. Numerical value:N: H: .Explanation: nitrogen is more electronegative than Hydrogen.
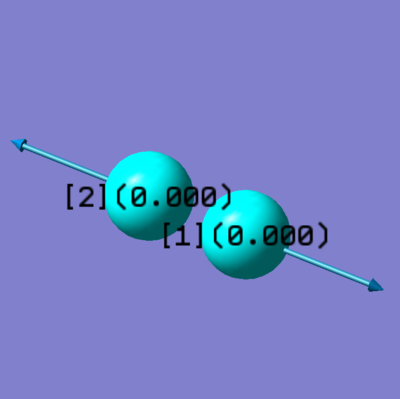
H2 molecules
- molecule name: hydrogen
- calculation method:RB3LYP
- basis set:6-31G(d,p)
- final energy E(RB3LYP): -1.17853936 a.u.
- the point group: C1
- RMS gradient:0.00000017 a.u.
- optimised H-H bond distance: 0.74279
test molecule |
The optimisation file is liked to here
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
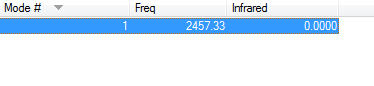
- Vibration:
- Charge Distribution:
- Charge on H-atoms: 0.000
N2 molecules
- molecule name: nitrogen
- calculation method:RB3LYP
- basis set:6-31G(d,p)
- final energy E(RB3LYP): -109.52412868 a.u.
- the point group: D*H
- RMS gradient:0.00000060 a.u.
- optimised N N bond distance: 1.10550
test molecule |
The optimisation file is liked to here
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400980D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad3
- Vibration:
- Charge Distribution:
- Charge on N-atoms: 0.000
Reaction energy for the reaction of N2 + 3H2 -> 2NH3
- E(NH3)= -56.55776873 a.u.
- 2*E(NH3)= -113.11553746 a.u.
- E(N2)= -109.52412868 a.u.
- E(H2)= -1.17853936 a.u.
- 3*E(H2)=-3.53561808 a.u.
- ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.0557907 a.u. = -146.47848285 kJ/mol
In summary, the ammonia product is more unstable because the forward reaction is exothermic.
BH3 molecules
- molecule name: trihydridoboron/borine
- calculation method:RB3LYP
- basis set:6-31G(d,p)
- final energy E(RB3LYP): -26.61532364 a.u.
- the point group: D3H
- RMS gradient:0.00000211 a.u.
- optimised B-H bond distance: 1.19232
- optimised H-B-H angle: 120.000
test molecule |
The optimisation file is liked to here
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000017 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-1.053682D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
- Vibration:
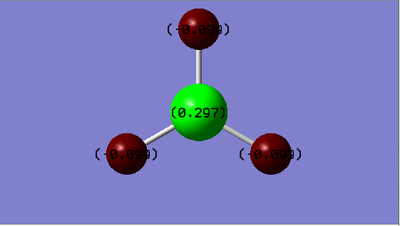
- Charge distribution:
- Charge on B-atom: 0.297
- Charge on H-atom:-0.099
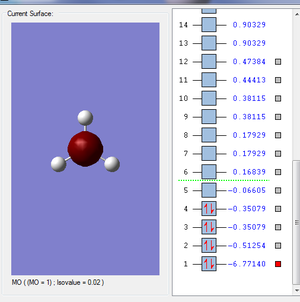
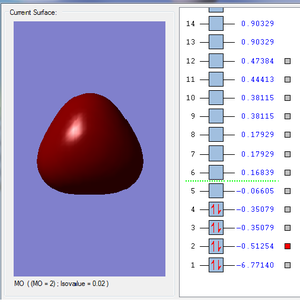
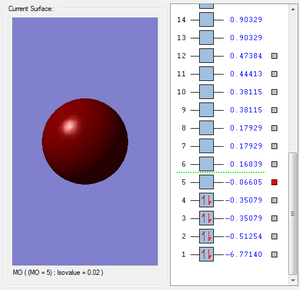
- Molecular Orbitals:
- Some examples of MOs:
- It is the MO with the lowest energy, it is the combination of the 1s orbital in B atom and the 1s orbital in H atoms.The orbital hardly overlap since 2s orbital is more likely to be attracted to the nuclear.
HCl molecules
- molecule name: hydrochloride
- calculation method:RB3LYP
- basis set:6-31G(d,p)
- final energy E(RB3LYP): -460.80077875 a.u.
- the point group: C*V
- RMS gradient:0.00005211 a.u.
- optimised B-H bond distance: 1.28599
test molecule |
The optimisation file is liked to here
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000090 0.000300 YES
Maximum Displacement 0.000139 0.001800 YES
RMS Displacement 0.000197 0.001200 YES
Predicted change in Energy=-1.256951D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.286 -DE/DX = 0.0001 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
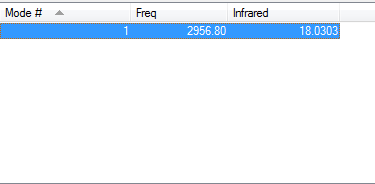
- Vibration:
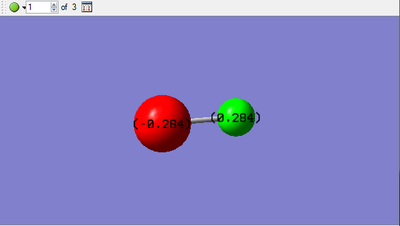
- Charge distribution:
- Charge on Cl-atom: -0.284
- Charge on H-atom:0.284