Rep:Mod:YkQ816
EX3
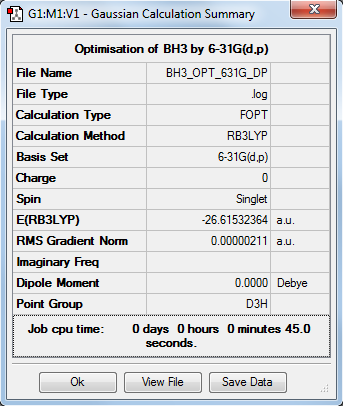
BH3
Optimisation
RB3LYP/6-31G(d,p)
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000003 0.000300 YES Maximum Displacement 0.000017 0.001800 YES RMS Displacement 0.000011 0.001200 YES
The molecular structure of BH3 |
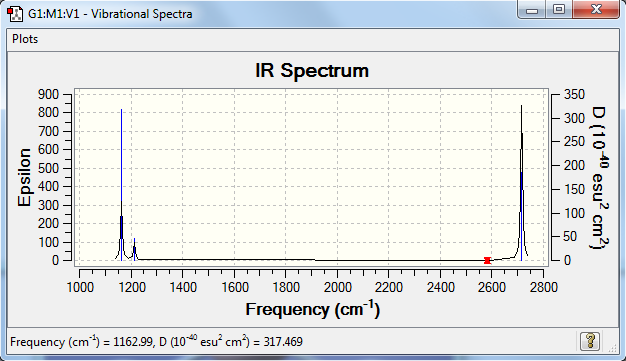
Frequencies
Frequency analysis log file YQ_BH3_FREQ.LOG
Low frequencies --- -1.1800 -1.0028 -0.0054 4.1927 11.0182 11.0637 Low frequencies --- 1162.9912 1213.1792 1213.1819
| wavenumber (cm-1) | Intensity (arbitrary units) | symmetry | IR active? | type |
| 1163 | 93 | A2 | yes | out-of-plane bend |
| 1213 | 14 | E | very slight | bend |
| 1213 | 14 | E | very slight | bend |
| 2582 | 0 | A1 | no | symmetric stretch |
| 2715 | 126 | E | yes | asymmetric stretch |
| 2715 | 126 | E | yes | asymmetric stretch |
The number of vibrations is smaller than the vibration peaks shown on the spectrum. In one hand, one of these vibrations is symmetric so there is no change of dipole moment. This type of vibration can not be detected by the IR spectrum. On the other hand, there are vibrations that are degenerate because the symmetry makes them have the same frequency of the change in dipole moment.
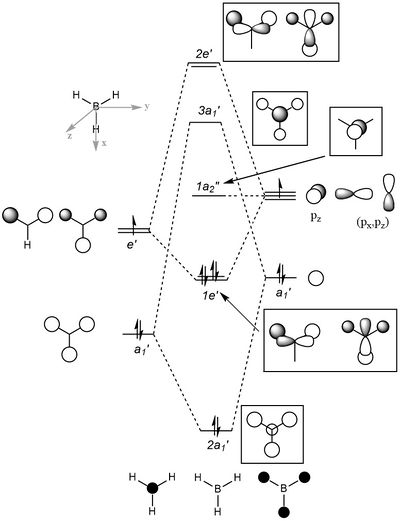
MOs
| Shape | Description |
|---|---|
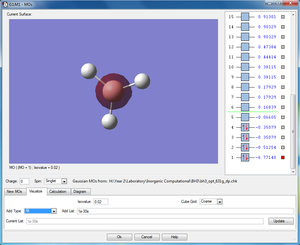 |
σ1s (Non-bonding Orbital) |
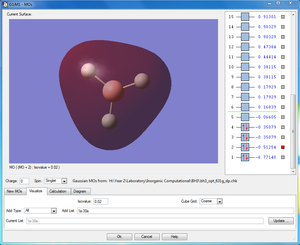 |
2a'x |
 |
1e'x |
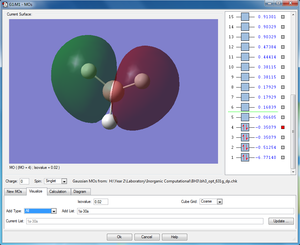 |
1e'y (HOMO) |
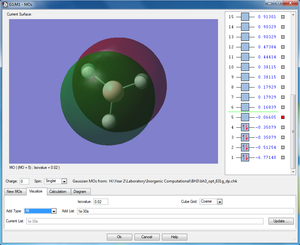 |
1a"2 (LUMO) |
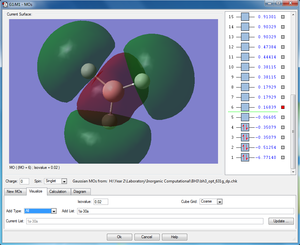 |
3a'1 |
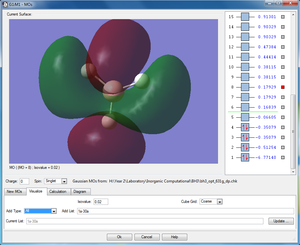 |
2e'x |
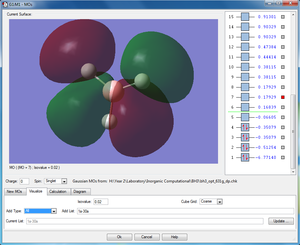 |
2e'y |
The relative distributions of the electrons of LCAOs and real MOs have no significant differences. However, LCAO is not able to determine the precise shape of the molecular orbitals. Nevertheless, it can still predict the general appearance of the molecular orbitals with minimum calculations.
NH3
Optimisation
RB3LYP/6-31G(d,p)
Item Value Threshold Converged? Maximum Force 0.000005 0.000450 YES RMS Force 0.000003 0.000300 YES Maximum Displacement 0.000010 0.001800 YES RMS Displacement 0.000007 0.001200 YES
The molecular structure of NH3 |
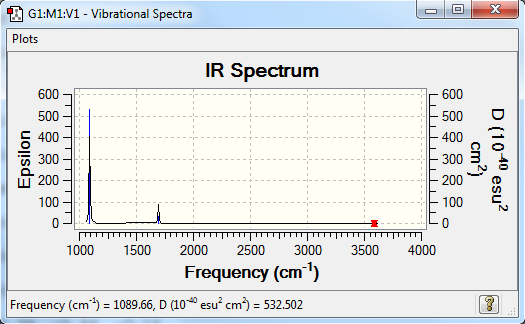
Frequencies
Frequency analysis log file YQ_NH3_6_OPT_FREQ.LOG
Low frequencies --- -11.6527 -11.6490 -0.0048 0.0332 0.1312 25.5724 Low frequencies --- 1089.6616 1694.1736 1694.1736
| wavenumber (cm-1) | Intensity (arbitrary units) | symmetry | IR active? | type |
| 1090 | 145 | A | yes | bend |
| 1694 | 14 | E | very slight | bend |
| 1694 | 14 | E | very slight | bend |
| 3461 | 1 | A | no | symmetric stretch |
| 3590 | 0 | E | no | asymmetric stretch |
| 3590 | 0 | E | no | asymmetric stretch |
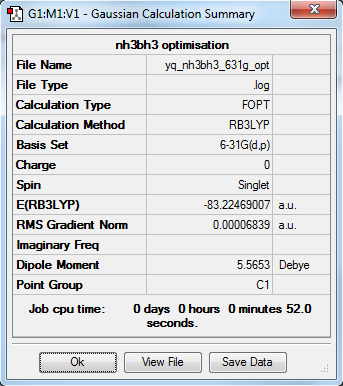
NH3BH3
Optimisation
RB3LYP/6-31G(d,p)
Item Value Threshold Converged? Maximum Force 0.000139 0.000450 YES RMS Force 0.000063 0.000300 YES Maximum Displacement 0.000771 0.001800 YES RMS Displacement 0.000338 0.001200 YES
The molecular structure of NH3BH3 |
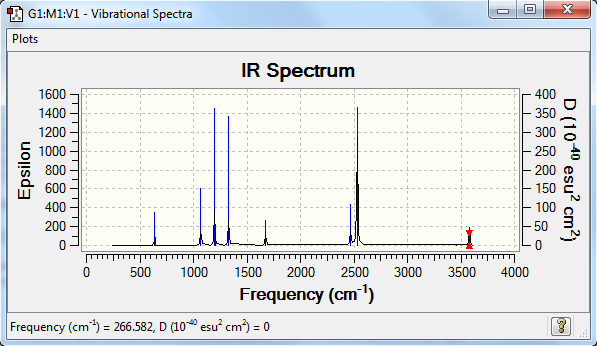
Frequecies
Frequency analysis log file YQ_NH3BH3_631G_OPT.LOG
Low frequencies --- -0.0011 -0.0004 0.0009 19.0336 23.6908 42.9675 Low frequencies --- 266.5859 632.3793 639.4649
| wavenumber (cm-1) | Intensity (arbitrary units) | symmetry | IR active? | type |
| 267 | 0 | A | no | torsion |
| 632 | 14 | A | very slight | symmetric stretch |
| 639 | 4 | A | no | bend |
| 640 | 4 | A | no | bend |
| 1069 | 41 | A | yes | bend |
| 1070 | 41 | A | yes | bend |
| 1197 | 109 | A | yes | bend |
| 1204 | 3 | A | no | bend |
| 1204 | 4 | A | no | bend |
| 1330 | 114 | A | yes | bend |
| 1676 | 28 | A | very slight | bend |
| 1676 | 28 | A | very slight | bend |
| 2470 | 67 | A | yes | symmetric stretch |
| 2530 | 231 | A | yes | asymmetric stretch |
| 2530 | 231 | A | yes | asymmetric stretch |
| 3462 | 3 | A | no | symmetric stretch |
| 3579 | 28 | A | very slight | asymmetric stretch |
| 3579 | 28 | A | very slight | asymmetric stretch |
Association Energy
Total energies of the reactants and the products
E(NH3)= -56.55777 a.u.
E(BH3)= -26.61532 a.u.
E(NH3BH3)= -83.22469 a.u.
The association energy of NH3BH3
ΔE = E(NH3BH3)-[E(NH3)+E(BH3)]
ΔE = (-83.22469)-[(-56.55777)+(-26.61532)]
ΔE = -0.0516 a.u.
ΔE = -135 kJ/mol
Conclusions
N-N bond strength is 159 kJ/mol which is a relatively weak bond. N-B bond strength is even weker. Therefore, the N-B bond is a weak bond.
Ng611 (talk) 23:40, 15 May 2018 (BST)Remember to cite your bond values (ideally from a textbook, databook, or paper).
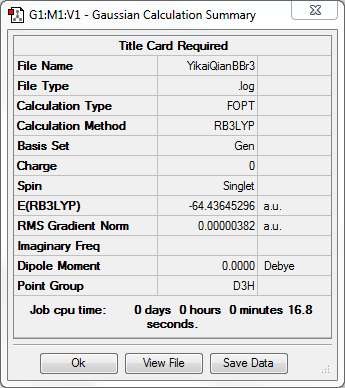
BBr3
Optimisation
RB3LYP/Gen
Item Value Threshold Converged? Maximum Force 0.000008 0.000450 YES RMS Force 0.000005 0.000300 YES Maximum Displacement 0.000036 0.001800 YES RMS Displacement 0.000023 0.001200 YES
The molecular structure of BBr3 |
DOI identifier
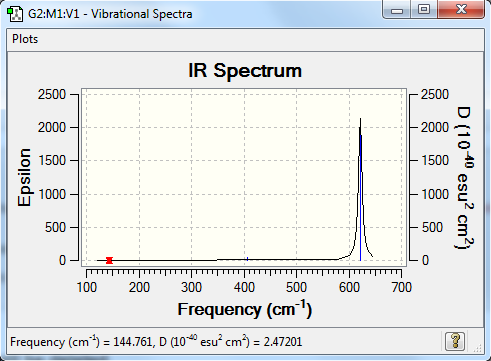
Frequencies
Frequency analysis log file YikaiQianBBr3_freq.log
Low frequencies --- -0.0116 -0.0065 -0.0003 49.9506 49.9506 50.0315 Low frequencies --- 144.7606 144.7640 215.6181
| wavenumber (cm-1) | Intensity (arbitrary units) | symmetry | IR active? | type |
| 145 | 0 | E' | no | in-plane bend |
| 145 | 0 | E' | no | in-plane bend |
| 216 | 0 | A1' | no | symmetric stretch |
| 407 | 5 | A2" | no | out-of-plane bend |
| 622 | 313 | E' | yes | in-plane bend |
| 622 | 313 | E' | yes | in-plane bend |
Lewis acids and bases
Structures of isomers
| Isomers | Symmetry |
| D2h | |
| C2h | |
| C2h | |
| C1 | |
| C2v |
Optimisations
Isomer1
RB3LYP/GEN
Item Value Threshold Converged? Maximum Force 0.000002 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000686 0.001800 YES RMS Displacement 0.000361 0.001200 YES
Isomer2
RB3LYP/GEN
Item Value Threshold Converged? Maximum Force 0.000022 0.000450 YES RMS Force 0.000008 0.000300 YES Maximum Displacement 0.001268 0.001800 YES RMS Displacement 0.000483 0.001200 YES
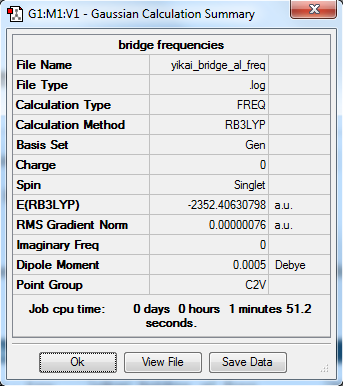
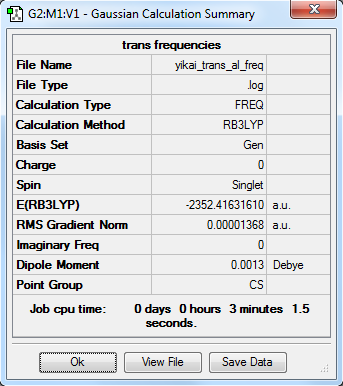
Energies
The total energy of isomer 1: E1 = -2352.40631 a.u. = -6176243 kJ/mol
The total energy of isomer 2: E2 = -2352.41632 a.u. = -6176269 kJ/mol
The relative energy of the two isomers: ΔEr = E2 - E1 = -26 kJ/mol
The relative energy of isomer 2 is more negative than that of isomer 1. This means that the stability of isomer with trans terminal Br is lower. This is due to the position of the bromine atoms that gives lowest repulsive force under this trans structure.
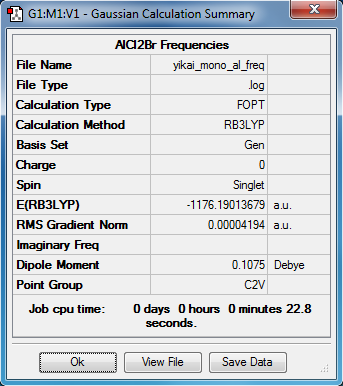
Monomer
Optimisation
The molecular structure of AlCl2Br |
RB3LYP/GEN
Item Value Threshold Converged? Maximum Force 0.000136 0.000450 YES RMS Force 0.000073 0.000300 YES Maximum Displacement 0.000906 0.001800 YES RMS Displacement 0.000543 0.001200 YES
Energies
The total energy of isomer 2: E2 = -2352.41632 a.u. = -6176269 kJ/mol
The total energy of monomer AlCl2Br: Emono = - 1176.19014 a.u. = - 3088087 kJ/mol
The dissociation energy of isomers 2: ΔEd = 2 * Emono - E2 = 95 kJ/mol
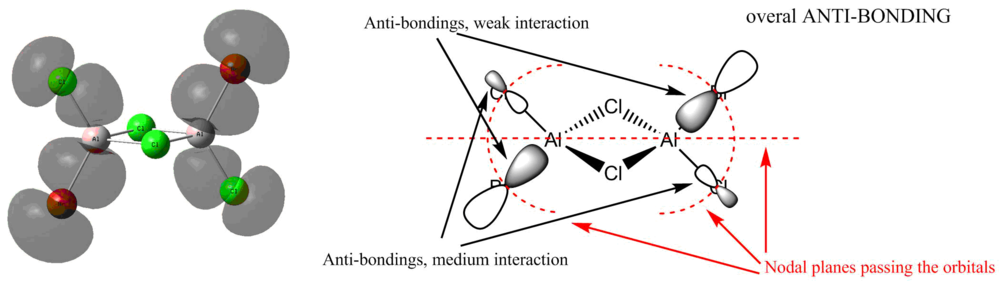
MOs of isomer2 by decreasing energies
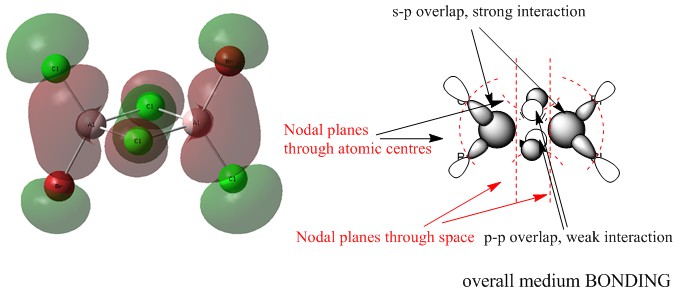
MO 45
Ng611 (talk) 23:43, 15 May 2018 (BST) Whilst this looks correct, I'm not able to check the parities of your orbitals as they're not coloured so it's hard for me to know for sure.
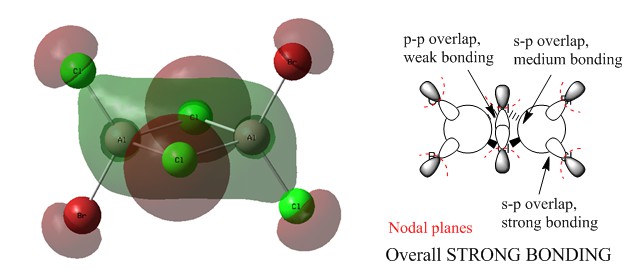
MO 41
MO 38
Ng611 (talk) 23:46, 15 May 2018 (BST) You've gotten the main interactions right but needed to specify whether they're through space/along bond and/or directed/not directed and what this says about the strength of the overall MO.
Ng611 (talk) 23:46, 15 May 2018 (BST) A good report. Good first section. You let yourself down in the second section by not giving the frequency files and low frequency mode calculations for any of the calculations. However, your MO analysis was good, well done.