Rep:Mod:YYTmod1
Year 3 Computational Lab
Module 1: The basic techniques of molecular mechanics and semi-empirical molecular orbital methods for structural and spectroscopic evaluations
Modelling Using Molecular Mechanics
- The Hydrogenation of Cyclopentadiene Dimer
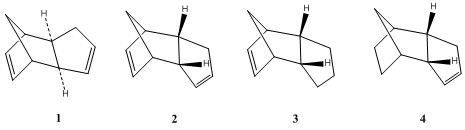
Stabilities of structure 1 and 2, 3 and 4 were compared by using MM2 force field option in ChemBio 3D. And the results were displayed in the table below.
| Structure | Stretch | Bend | Stretch-Bend | Torsion | Non-1,4 Van de Waal's | 1,4 Van de Waal's | Dipole/Dipole | Total Energy |
|---|---|---|---|---|---|---|---|---|
| 1 | 1.2500 | 20.8490 | -0.8356 | 9.5109 | -1.5434 | 4.3191 | 0.4475 | 33.9975 kcal/mol |
| 2 | 1.2850 | 20.5783 | -0.8382 | 7.6559 | -1.4167 | 4.2346 | 0.3775 | 31.8765 kcal/mol |
| 3 | 1.2395 | 18.7708 | -0.7514 | 12.7266 | -1.3375 | 6.0492 | 0.1632 | 36.8603 kcal/mol |
| 4 | 1.1300 | 13.0132 | -0.5653 | 12.4121 | -1.3246 | 4.4411 | 0.1410 | 29.2475 kcal/mol |
The stretching energy describes positions of atoms related to the equilibrium positions, whereas the bending energy depends on angles between three atoms in the molecule. Since the bond angles might vary with the change of bond length, the stretch-bend term was also calculated. Ideally, the length, angle and torsional term involved in steric energy tend to zero, and positive values lead to instability.
As shown above in the table, the total steric energy of 1 is slightly larger than that of 2, which means, structure 2 is relatively more stable, though it has more positive energy in stretching and torsion.
Similarly, 4 can be regarded as a more stable structure than 3 due to its much lower steric energy. That is, 4 should be the prefered product according to the data above. The dipole/dipole contribution of the four structures were all small because there was no typical hydrogen bond (e.g. hydrogen bonding between hydrogen and oxygen). But 3 and 4 had even weaker dipole/dipole interaction since they had one less electron rich C=C double bonds than 1 and 2. One the other hand, the larger 1,4 van der waal term of 3 indicated that some hydrogen atoms might be closer to each other than those in 4. Also, the large bending term reflected the dramatic changes in bond angles.
- Stereochemistry and Reactivity of an Intermediate in the Synthesis of Taxol
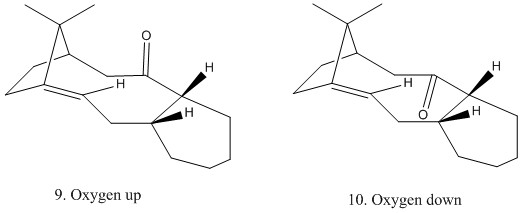
Similarly, the stabilities if these immediates were compared using MM2 in ChemBio 3D. And the results were as below.
| Intermediate | MM2 | MMFF94 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Stretch | Bend | Stretch-Bend | Torsion | Non-1,4 Van de Waal's | 1,4 Van de Waal's | Dipole/Dipole | Total Energy | ||
| 9(click to view jmol) | 2.9491 | 17.2140 | 0.5012 | 21.2872 | -1.4216 | 14.5051 | -1.7305 | 53.3045 kcal/mol | 82.225 kcal/mol |
| 10(click to view jmol) | 2.8324 | 12.9762 | 0.3901 | 22.9833 | -1.7561 | 14.1197 | -2.0324 | 49.5132 kcal/mol | 75.160 kcal/mol |
As shown above, intermediate 10 is preferred due to smaller steric energy. Every energy term was similar for both intermediate structures, except bending energy, which was slightly larger for 9. The total energies calculated using MMFF94 method were both larger than those computed using MM2, though the one of 10 was lower than that of 9, which was the same as the MM2 result.
These intermediates could be easily produced through [3,3]-sigmatropic reaction called oxy-cope rearrangement.
Since it is accompanied by significant strain energy release, this transformation is considered as generally irreversible due to the big energy gap between the starting material and the product. However, the reaction could be reversible in this case as the two diene termini keep close to each other, even though the C=C bond decomposes slowly.
References
1. S.W. Elmore and L. Paquette, Tetrahedron Letters, 1991, 319 (DOI:10.1016/S0040-4039(00)92617-0 )
2. See J.G. Vinter and H. M. R.Holffman, J. Am. Chem. Soc, 1974, 96, 5466 (DOI:10.1021/ja00824a025 )
Modelling Using Semi-empirical Molecular Orbital Theory
- Rgioselective Addition of Dichlorocarbene
In this case, MOPAC/PM6 method was used to calculate molecular orbitals in order to explore the selectivity of reactions start from the folowing molecule:
MM2 method was used first, and then the MOPAC calculation. And finally the Gaussian computation was run to find out the vibrational data. Computational results were displayed as below:
| Optimized Structure | MM2 | MOPAC/PM6 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Stretch | Bend | Stretch-Bend | Torsion | Non-1,4 Van de Waal's | 1,4 Van de Waal's | Dipole/Dipole | Total Energy | Heat of Formation | |
| Diene (click to view jmol) | 0.6195 | 4.7370 | 0.0401 | 7.6591 | -1.0672 | 5.7937 | 0.1123 | 17.8945 kcal/mol | 19.74040 kcal/mol |
| Monoene (click to view jmol) | 0.8973 | 4.6915 | 0.0138 | 10.7584 | -1.0664 | 6.9712 | 0.0708 | 22.3365 kcal/mol | -2.43088 kcal/mol |
|
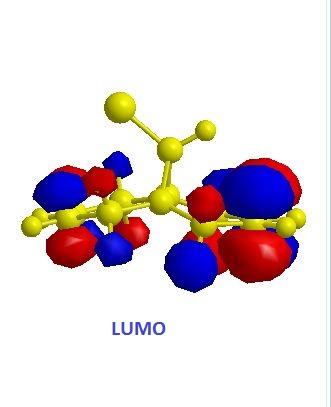 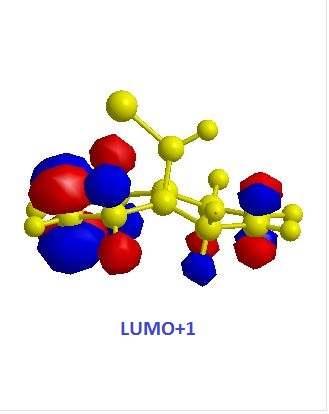 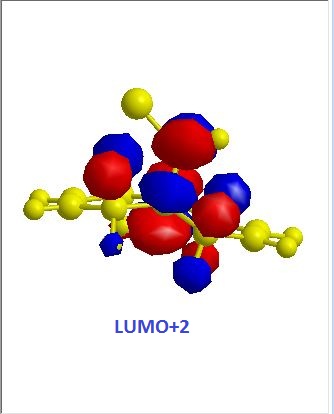
|
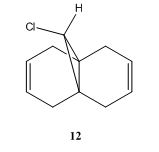
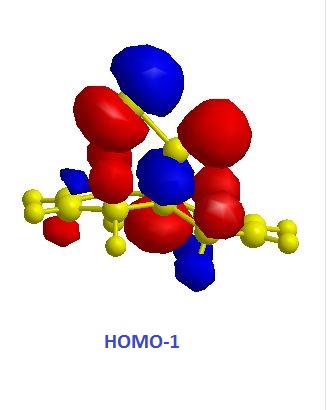
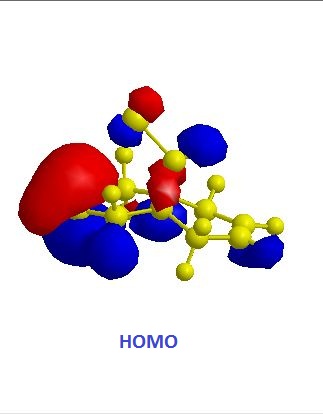
The exo alkene is more stable due to antiperiplanar overlap between its π-orbital and the σ anti-bonding orbital of C-Cl. And this also results in more nucleophilic endo C=C double bond. The molecular orbitals of 12 were obtained by using MOPAC/PM6 method, and the results matched the literature ones.(DOI:10.1039/P29920000447 ) In the HOMO diagram shown above, the orbital on the endo alkene was the most significant, which proved its larger reactivity.
| Diene (click to view spectrum) | 1736.93cm-1(C=C stretch), 1757.41cm-1(C=C stretch), 3009.26cm-1(=C-H stretch), 3028.57(=C-H stretch), 689.60cm-1(C-Cl stretch) |
| Monoene (click to view spectrum) | 1758.05cm-1(C=C stretch), 3030.91cm-1(=C-H stretch), 673.90cm-1(C-Cl stretch) |
The two spectra were generally similar, except the alkene stretches. The IR spectrum of diene showed double peaks for C=C stretch whereas that of monoene had only one, since 12 had one more C=C double bond than its monohydrogenated derivative had. Obviously, the exo alkene had smaller frequency which again proved it was more stable than the endo one.
References
1. B. Halton, R. oeses and H. S. Rzepa., J. Chem. Soc., Perkin Trans 2, 1992, 447 (DOI:10.1039/P29920000447 )
- Monosaccharide Chemistry: glycosidation
Methyl group was used as the R group in this calculation since it has only four atoms and fewer angles and bonds involved in the steric energy than other alkyl groups. MM2 method was preferred in this case as it was fast and systems compared had no breaking or forming bonds.
Both MM2 and MOPAC/PM6 were run, and the results were displayed as below
| Structure | MM2 | MOPAC/PM6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Stretch | Bend | Stretch-Bend | Torsion | Non-1,4 Van de Waal's | 1,4 Van de Waal's | Charge/Dipole | Dipole/Dipole | Total Energy | Heat of Formation | |
| A | 2.3904 | 10.1639 | 0.8999 | 0.9232 | -2.6100 | 18.6712 | -3.1310 | 4.0057 | 31.3133 kcal/mol | -77.38771 kcal/mol |
| A' | 2.3817 | 13.3282 | 1.0052 | 0.7568 | -2.4934 | 18.1786 | -0.4410 | 8.0355 | 40.7516 kcal/mol | -69.69350 kcal/mol |
| B | 2.4771 | 10.2391 | 0.8773 | 1.1447 | -2.2308 | 18.7249 | -3.9381 | 4.6223 | 31.9164 kcal/mol | -77.31141 kcal/mol |
| B' | 2.6793 | 14.6942 | 1.1162 | 0.1851 | -1.5201 | 17.9199 | 3.5813 | 7.5381 | 46.1940 kcal/mol | -68.23032 kcal/mol |
| C | 2.7319 | 17.3219 | 0.7908 | 8.2174 | -2.6223 | 19.3420 | 2.0574 | -1.7293 | 46.1097 kcal/mol | -66.84415 kcal/mol |
| D | 2.0317 | 15.4353 | 0.6903 | 6.8576 | -4.0343 | 17.7646 | 2.7133 | -0.2945 | 41.1460 kcal/mol | -85.93973 kcal/mol |
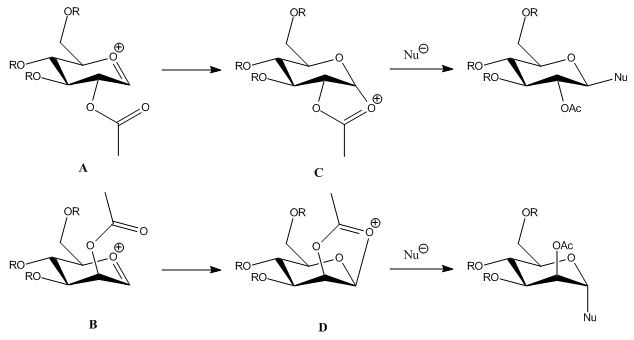
Conformers with the acyl group pointing to opposite direction was found for structure A and B, but the steric energy tended to be lower when the oxygen of acyl group approached the carbon of the double bond in the oxonium cation (structure A and B) as the result of electrostatic attraction between these two atoms. Comparing A and B, their steric energy were close to eachother, which indicated the similar stabilities of them. However, similar conformers could not be found for C and D since the free rotation along the C-O bond of acyl group was trapped due to the newly formed C-O bond. C seemed to be more stable since its had both lower steric energy and heat of formation. Tables below shows the bond lengths and bond angles of both structure optimized by the two methods which were calculated by ChemBio3D for comparison. The results obtained using both methods were very similar, but that of MM2 seemed to be slightly closer to the optimal value. And that proved MM2 the more suitable method in this task.
|
|
|
| ||||
|
|
|
|
| |||
|
|
|
|
| |||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| |||||
|
|
|
|
| |||
|
|
|
|
| |||
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
References
1. D. M. Whitfield, T. Nukada, Carbohydr. Res., 2007, 342, 1291 (DOI:10.1016/j.carres.2007.03.030 )
Structure based Mini project using DFT-based Molecular orbital methods
Bromination of 1-(2-Pyridyl)-2-propen-1-ol(DOI:10.1021/jo201830b )
- Background
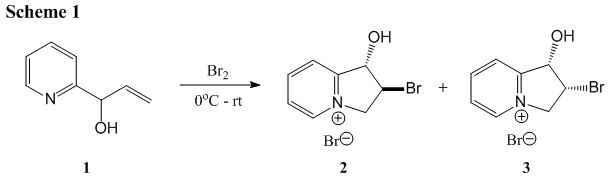
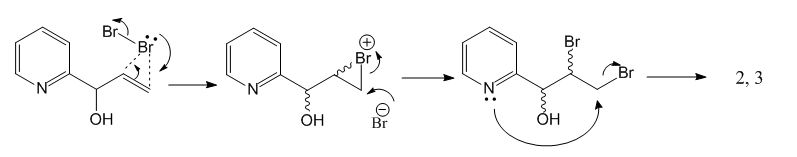
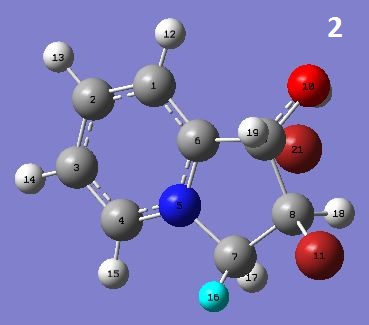
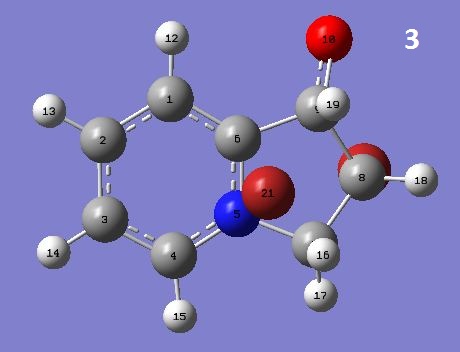
This is the first step of 1,2-dihydroxyindolizidines synthesis from 1-(2-pyridyl)-2-propen-1-ol, during which a pair of diastereomers, noted as 2 and 3, are formed. Below is the mechanism of this reaction.
- Procedure
For diastereomers, H NMR can be used to distinguish the two structure since they have different coupling constans. To study these two molecules, steric energy was firstly minimised using MM2 method in ChemBio3D, followed by computing Gaussian optimised structure, and finally 13C NMR spectra as well as H-H coupling constants were calculated by SCAN and Janocchio respectively.
- Results and Discussion
| Diastereomer | Structure | method | Stretch | Bend | Stretch-Bend | Torsion | Non-1,4 Van de Waal's | 1,4 Van de Waal's | Charge/Dipole | Dipole/Dipole | Total Energy |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 0.4924 | 4.9221 | -0.0206 | -0.6962 | 3.2589 | 5.9525 | -73.7736 | -15.8347 | 1.2080 | -74.4912 kcal/mol | |
| 3 | 0.5989 | 5.3749 | -0.0665 | -0.1433 | 2.1316 | 6.9154 | -73.1238 | -11.7016 | 0.8690 | -69.1455 kcal/mol |
| Diastereomer | Calculated Spectrum | Literature Spectrum(DOI:10.1021/jo201830b ) |
|---|---|---|
| 2 | δ136.2, δ128.8, δ110.4, δ75.1, δ74.1, δ60.4, δ55.1, δ36.7 (Click to view spectrum) (DOI:10042/to-9524 ) | δ156.6(s), δ147.0(d), δ141.8(d), δ127.6(d), δ124.5(d), δ78.95(d), δ62.7(t), δ46.8(d) |
| 3 | δ129.1, δ126.6, δ119.7, δ73.8, δ72.8, δ58.3, δ54.1, δ37.8 (Click to view spectrum) (DOI:10042/to-9525 ) | δ157.1(s), δ146.8(d), δ142.2(d), δ127.2(d), δ124.4(d), δ73.4(d), δ64.2(t), δ55.1(d) |
From the MM2 results, 2 seemed to be more stable as it has lower steric energy. The Calculated 13C NMR signals were generally smaller than the literature ones. Also, the splitting of peaks were not shown in the output. However, the number of signals and the relative positions roughly matched those of literature spectra.
Diastereomers have different NMR H-H coupling constants. But it cannot be confirmed which is which unless comparing it to that of known structures. In this case, the JH19-H18 of 2 was calculated as 3.9037ppm, whereas that of 3 was 4.7951ppm, and the difference was large enough to differ the two diastereomers.
- References
Synthesis of 1,2-Dihydroxyindolizidines from 1-(2-Pyridyl)-2-propen-1-ol, Donatella Giomi, 2011 (DOI:10.1021/jo201830b )