Rep:Mod:YYL0508
Loh Yong Yao
CID: 00605583
BH3 Optimisation
3-21G Optimisation
Link to BH3 3-21G Optimisation Log File
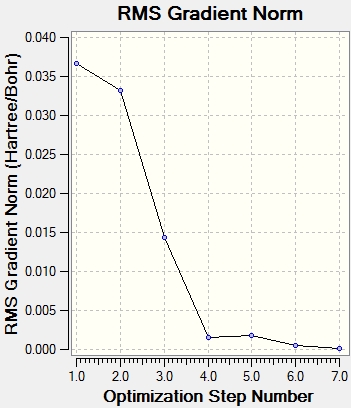
The process of the optimisation can be observed from the minimisation of total energy and gradient as seen in the graphs below:

The summary of the optimisation is as follows:
File type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 3-21G
Final Energy:-26.46226429 a.u.
Gradient: 0.00008851 a.u.
Dipole Moment: 0.00 D
Point Group: D3h
Job Time: 17.0 s
Item Value Threshold Converged?
Maximum Force 0.000220 0.000450 YES
RMS Force 0.000106 0.000300 YES
Maximum Displacement 0.000940 0.001800 YES
RMS Displacement 0.000447 0.001200 YES
Predicted change in Energy=-1.672479D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1948 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1944 -DE/DX = -0.0001 !
! R3 R(1,4) 1.1947 -DE/DX = -0.0002 !
! A1 A(2,1,3) 119.986 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0157 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9983 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
6-31g(d,p) Optimisation
Link to BH3 6-31g(d,p) Optimisation Log File
The summary of the optimisation is as follows:
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Energy: -26.61532372 a.u.
Gradient: 0.00001746 a.u.
Dipole Moment: 0.00 D
Point Group: D3h
Job Time: 6.0 s
Item Value Threshold Converged?
Maximum Force 0.000019 0.000450 YES
RMS Force 0.000013 0.000300 YES
Maximum Displacement 0.000090 0.001800 YES
RMS Displacement 0.000047 0.001200 YES
Predicted change in Energy=-2.991560D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1924 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0126 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9883 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9991 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Summary
Optimised BH3 Bond Distance: 1.19 Å (comparable to lit value = 1.19 Å)[1]
Optimised H-B-H Bond Angle: 120.0 ° (consistent with D3h Point Group)
Energy for 3-21G Optimised Structure: -26.46226429 a.u.
Energy for 6-31G(d,p) Optimised Structure: -26.61532372 a.u.
GaBr3 Optimisation
LanL2DZ Optimisation (calculated on HPC)
Link to GaBr3 LanL2DZ Optimisation Log File
The summary of the optimisation is as follows:
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: LANL2DZ
Energy: -41.70082783 a.u.
Gradient: 0.00000016 a.u.
Dipole Moment: 0.00 D
Point Group: D3h
Job Time: 13.9 s
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000003 0.001800 YES
RMS Displacement 0.000002 0.001200 YES
Predicted change in Energy=-1.282685D-12
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.3502 -DE/DX = 0.0 !
! R2 R(1,3) 2.3502 -DE/DX = 0.0 !
! R3 R(1,4) 2.3502 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Optimised GaBr3 Bond Distance: 2.35 Å (comparable to lit value = 2.3525 Å)
[1]
Optimised Br-Ga-Br Bond Angle: 120.0 ° (consistent with D3h Point Group)
Energy for LanL2DZ Optimised Structure: -41.70082783 a.u.
BBr3 Optimisation
6-31G(d,p) + LanL2DZ Optimisation
Link to BBr3 6-31G(d,p) + LanL2DZ Optimisation Log File
The summary of the optimisation is as follows:
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: GEN
Energy: -64.43644917 a.u.
Gradient: 0.00001229 a.u.
Dipole Moment: 0.00 D
Point Group: D3h
Job Time: 20.7 s
Item Value Threshold Converged?
Maximum Force 0.000035 0.000450 YES
RMS Force 0.000020 0.000300 YES
Maximum Displacement 0.000207 0.001800 YES
RMS Displacement 0.000125 0.001200 YES
Predicted change in Energy=-5.432275D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.9339 -DE/DX = 0.0 !
! A1 A(2,1,3) 119.9878 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0008 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0113 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Optimised B-Br Bond Distance: 1.93 Å (comparable to lit value = 1.893)[1]
Optimised Br-B-Br Bond Angle: 120.0 ° (consistent with D3h Point Group)
Energy for LanL2DZ Optimised Structure: -64.43644917 a.u.
Analysis of Results
Comparison between BH3, GaBr3 and BBr3
| Molecule | Bond Length (Å) |
|---|---|
H and Br are similar in that they both require one more electron to attain a full valence shell. Comparing H and Br as ligands with B as the central atom, the bond length of central atom to ligand increases from 1.19 Å to 1.93 Å. This is probably due to the fact that B is in the 2nd row of the periodic table and has much better orbital overlap with H than with Br, which is in the 4th row. This is because there is a better match in orbital size between atoms in the same row of the periodic table. Also, the orbitals are more diffuse down the group and overlap less effectively. Also, Br is a much larger atom as compared to H, hence, the internuclear distance between B and Br is larger than that between B and H. Therefore, the B-H bond is shorter than the B-Br bond.
Comparing B and Ga as the central atom, they are both in group 3, but Ga is 2 rows below B, and is in the same row as the ligand Br. Hence, Ga is a much larger atom as compared to B, hence the Ga-Br bond length, which is the internuclear distance, is larger as compared to the internuclear distance between a small B atom and Br. Also, orbitals are more diffused down the group, hence, there is less effective overlap and the bond is weaker and longer. This outweighs the fact that there is better orbital size match between Ga and Br.
A bond can loosely be defined as the overlap of atomic orbitals which allows for the delocalisation of electrons between two nuclei, lowering the energy of the electrons. In some structures, this overlap is very small, possibly due to the distance between the central atom and the ligand and hence, stabilisation is very small. However, there is still a bond between the atoms, albeit a weak one. GaussView draws bonds based on a distance critera, so the fact that GaussView hasn't drawn bonds doesn't mean they are not there. Rather, it just means that the bond is longer than the pre-set distance criteria for a covalent bond.
This is shown in the 7 different strutures obtained when BH3 was optimised using GaussView. Initially, the H atoms are too far apart from the central B atom, and no bond is drawn between the B and H atoms. However, as the optimisation went on, a bond was eventually formed in the later structures as the H atoms were moved closer to the central B atoms and satisfied the distance criteria in GaussView. This also coincides with an increase in orbital overlap and stabilisation of the electrons.
Further Analysis of BH3
Frequency Calculations for BH3
Link to Log File for Frequency Calculations of BH3
The summary of the frequency calculation is as follows:
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Energy: -26.61532364 a.u.
Gradient: 0.00000003 a.u.
Dipole Moment: 0.00 D
Point Group: D3h
Job Time: 6.0 s
Low frequencies --- -9.3212 -9.3057 -0.0697 0.0001 0.5380 2.6444 Low frequencies --- 1162.9905 1213.1497 1213.1499
Item Value Threshold Converged?
Maximum Force 0.000000 0.000002 YES
RMS Force 0.000000 0.000001 YES
Maximum Displacement 0.000000 0.000006 YES
RMS Displacement 0.000000 0.000004 YES
Predicted change in Energy=-1.929123D-14
Optimization completed.
-- Stationary point found.
Vibrations
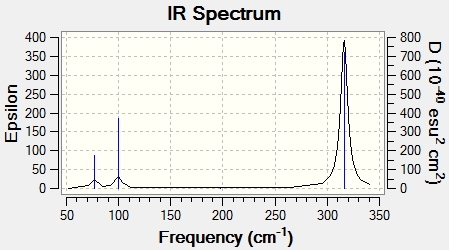
IR Spectrum

There are 6 vibrational modes, however, one of them has 0 intensity, while there are two pairs of degenerate vibrational modes. The mode with 0 intensity is the symmetrical stretch mode, and it has no intensity due to the fact that the stretching does not change the dipole moment of the molecule. There must be a change in dipole moment in order for the vibrational mode to show up on the IR spectrum. Hence, only 3 vibrational modes are observed in the IR spectrum.
Further Analysis of GaBr3
Frequency calculations for GaBr3
Link to Log File for Frequency calculation for GaBr3
Link to Dspace
The summary of the frequency calculation is as follows:
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: LanL2DZ
Energy: -41.70082783 a.u.
Gradient: 0.00000011 a.u.
Dipole Moment: 0.00 D
Point Group: D3h
Job Time: 11.3 s
Low frequencies --- -0.5252 -0.5247 -0.0024 -0.0010 0.0235 1.2010 Low frequencies --- 76.3744 76.3753 99.6982
Lowest real normal mode is 76.3744 cm-1
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000002 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-6.142863D-13
Optimization completed.
-- Stationary point found.
IR Spectrum
Comparison between the vibrations of BH3 and GaBr3
The atoms move in the same direction at the same time. This is a bending mode. |
|||
The atoms move in the same direction at the same time. This is a bending mode. |
|||
This is a bending mode. |
|||
This is a symmetric stretching mode. |
|||
This is an asymmetric stretching mode. |
|||
However, only two of them are synchronized. This is an asymmetric stretching mode. |
The large difference in values of the frequencies for BH3 compared to GaBr3 indicate a large difference in bond strength. Comparing the frequencies and the vibrational modes of the molecules, there has been a reordering of modes for GaBr3 relative to BH3. The spectra for both molecules are similar in that both have 3 signals, and this is because both have a vibrational mode that has 0 intensity as well as two pairs of degenerate modes.
A2" and E' modes are associated with wagging and scissoring respectively, hence they are of lower energies, while A1' and the other E' modes are associated with symmetrical stretching and asymmetrical stretching respectively, and stretching modes of vibrations are of higher energies.
The frequency analysis is essentially the second derivative of the potential energy surface, while the optimisation when the first derivative of the potential energy surface is zero (i.e. a turning point). Hence the same method and basis set must be used for both the optimisation and frequency analysis to ensure that the frequency analysis is conducted for the same potential energy surface that optimisation was carried out on.
The purpose of carrying out a frequency calculation is twofold. Firstly, it provides predicted IR and Raman modes for comparison with experimental values. Secondly and more importantly, it tells us if the optimisation that we have performed coincides with a minimum point on the potential energy surface. If all the frequencies are positive, then the optimisation is that of the minimum energy. If all the frequencies are negative, then the optimisation is that of a transition state.
The first 6 low frequencies represent the translational and rotational frequencies of the molecule, while the last 3 frequencies represent the vibrational frequencies of the molecule. This is the origin of the 3N - 6 formula for the number of vibrational frequencies of a non-linear molecule.
MO analysis
BH3
Link to Log File
Dspace Link
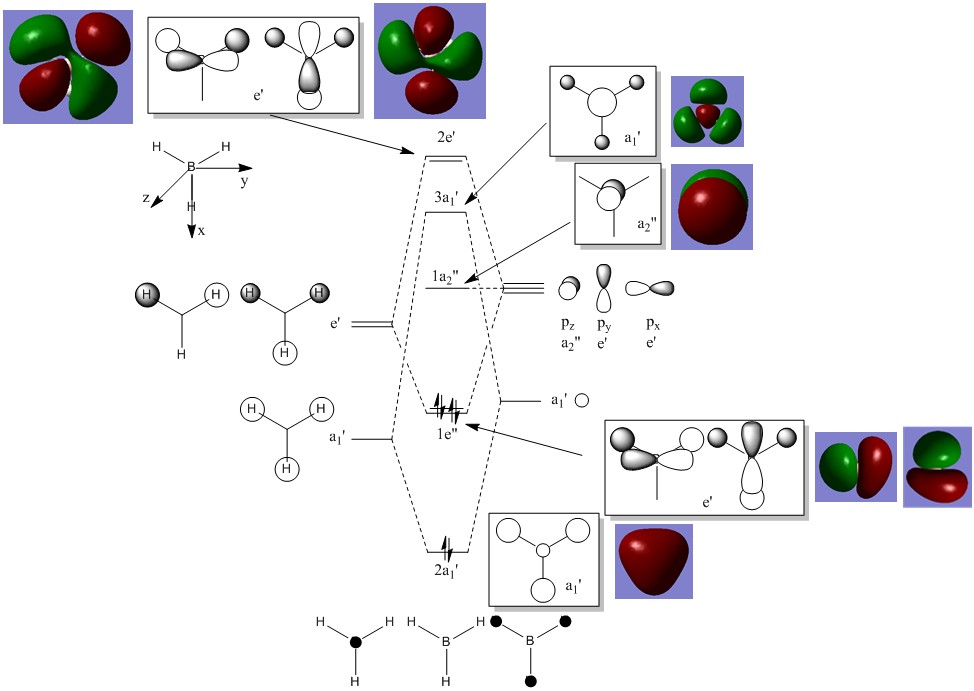
There are no real significant differences between the real and LCAO MOs, hence, qualitative MO theory can be considered to be reasonably accurate and really useful.
NBO Analysis
6-31G(d,p) Optimisation
Link to NH3 6-31G (d,p) Optimisation Log File
The summary of the optimisation is as follows:
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Energy: -56.55776854 a.u.
Gradient: 0.00003886 a.u.
Dipole Moment: 1.85 D
Point Group: C3v
Job Time: 11.0 s
Item Value Threshold Converged?
Maximum Force 0.000077 0.000450 YES
RMS Force 0.000044 0.000300 YES
Maximum Displacement 0.000174 0.001800 YES
RMS Displacement 0.000112 0.001200 YES
Predicted change in Energy=-1.601487D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = -0.0001 !
! R2 R(1,3) 1.018 -DE/DX = -0.0001 !
! R3 R(1,4) 1.0181 -DE/DX = -0.0001 !
! A1 A(2,1,3) 105.7546 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7438 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.743 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8739 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Frequency
Link to NH3 Frequency Log File
Link to Dspace
The summary of the frequency calculation is as follows:
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Energy: -56.55776872 a.u.
Gradient: 0.00000029 a.u.
Dipole Moment: 1.85 D
Point Group: D3h
Job Time: 20.7 s
Low frequencies --- -7.8411 -6.1113 -3.4648 -0.0019 -0.0018 -0.0008 Low frequencies --- 1089.3523 1693.9243 1693.9288
Item Value Threshold Converged?
Maximum Force 0.000000 0.000002 YES
RMS Force 0.000000 0.000001 YES
Maximum Displacement 0.000003 0.000006 YES
RMS Displacement 0.000001 0.000004 YES
Predicted change in Energy=-1.142027D-12
Optimization completed.
-- Stationary point found.
MO
Link to NH3 MO Log File
DSpace Link to population analysis
NBO

Charge Range: -1.125 to 1.125
Charge on N: -1.125
Charge on each H: +0.375
Association Energies
Optimisation
Link to NH3BH3 6-31G(d,p) Optimisation Log File
The summary of the optimisation is as follows:
File Type: .log
Calculation Type: FOPT
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Energy: -83.22468911 a.u.
Gradient: 0.00000009 a.u.
Dipole Moment: 5.56 D
Point Group: C1
Job Time: 49.0 s
Item Value Threshold Converged?
Maximum Force 0.000000 0.000002 YES
RMS Force 0.000000 0.000001 YES
Maximum Displacement 0.000000 0.000006 YES
RMS Displacement 0.000000 0.000004 YES
Predicted change in Energy=-1.536215D-14
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.0185 -DE/DX = 0.0 !
! R2 R(2,8) 1.0185 -DE/DX = 0.0 !
! R3 R(3,8) 1.0185 -DE/DX = 0.0 !
! R4 R(4,7) 1.2098 -DE/DX = 0.0 !
! R5 R(5,7) 1.2098 -DE/DX = 0.0 !
! R6 R(6,7) 1.2098 -DE/DX = 0.0 !
! R7 R(7,8) 1.6677 -DE/DX = 0.0 !
! A1 A(4,7,5) 113.8738 -DE/DX = 0.0 !
! A2 A(4,7,6) 113.8738 -DE/DX = 0.0 !
! A3 A(4,7,8) 104.5974 -DE/DX = 0.0 !
! A4 A(5,7,6) 113.8738 -DE/DX = 0.0 !
! A5 A(5,7,8) 104.5975 -DE/DX = 0.0 !
! A6 A(6,7,8) 104.5975 -DE/DX = 0.0 !
! A7 A(1,8,2) 107.876 -DE/DX = 0.0 !
! A8 A(1,8,3) 107.876 -DE/DX = 0.0 !
! A9 A(1,8,7) 111.0228 -DE/DX = 0.0 !
! A10 A(2,8,3) 107.8758 -DE/DX = 0.0 !
! A11 A(2,8,7) 111.023 -DE/DX = 0.0 !
! A12 A(3,8,7) 111.023 -DE/DX = 0.0 !
! D1 D(4,7,8,1) -180.0 -DE/DX = 0.0 !
! D2 D(4,7,8,2) -60.0 -DE/DX = 0.0 !
! D3 D(4,7,8,3) 60.0 -DE/DX = 0.0 !
! D4 D(5,7,8,1) -60.0 -DE/DX = 0.0 !
! D5 D(5,7,8,2) 60.0 -DE/DX = 0.0 !
! D6 D(5,7,8,3) 180.0 -DE/DX = 0.0 !
! D7 D(6,7,8,1) 60.0 -DE/DX = 0.0 !
! D8 D(6,7,8,2) -180.0 -DE/DX = 0.0 !
! D9 D(6,7,8,3) -60.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Frequency
Link to NH3BH3 Frequency Log File
Dspace Link
The summary of the frequency calculation is as follows:
File Type: .log
Calculation Type: FREQ
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Energy: -83.22468911 a.u.
Gradient: 0.00000023 a.u.
Dipole Moment: 5.56 D
Point Group: C1
Job Time: 104.8 s
Low frequencies --- -2.9083 -1.3024 -0.0008 0.0007 0.0010 3.6217 Low frequencies --- 263.3601 632.9766 638.4443
Item Value Threshold Converged?
Maximum Force 0.000000 0.000002 YES
RMS Force 0.000000 0.000001 YES
Maximum Displacement 0.000001 0.000006 YES
RMS Displacement 0.000001 0.000004 YES
Predicted change in Energy=-8.207670D-13
Optimization completed.
-- Stationary point found.
Calculation
The dissociation energy can be calculated as follows:
E(NH3) = -56.55776854 a.u.
E(BH3) = -26.61532372 a.u.
E(NH3BH3) = -83.22468911 a.u
ΔE = E(NH3BH3) - [E(NH3) + E(BH3)] = -82.22468911 - (-56.55776854 + -26.61532372) = -0.05159685 a.u. = -135.5 kJ mol-1
This gives the association energy, hence the dissociation energy is +135.5 kJ mol-1
This is a reasonable value for as typical dissociation energies are in the range of 100 to 500 kJ mol-1.
Additionally, I would consider the association between NH3 and BH3 to be a weak one as it is a dative covalent bond, hence, a relatively low value of +135.5 is reflective of a relatively low dissociation energy when compared to typical values, which indicates that the bond is weak.
References
<references> [1]