Rep:Mod:YW7216
NH3
Calculation Method RB3LYP
Basis set 6-31G(d.p)
Final energy E(RB3LYP) -56.44397188 a.u.
Point group C3V
RMS gradient norm 0.00000485 a.u.
NH3 |
N-H bond length 1.30Å
H-N-H bond angle 109.471°
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
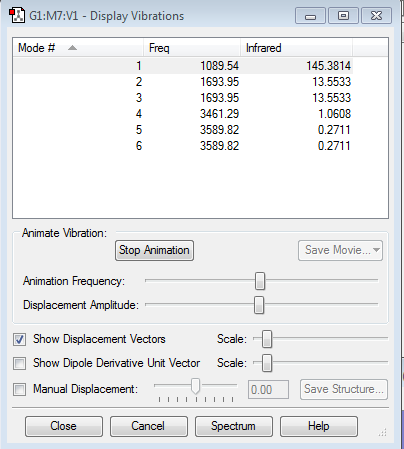
1.3 modes are expexted from 3N-6
2.mode5 and mode6 are degenerate, mode2 and mode3 are also degenerate.
3.mode1, 2 and 3 are bending vibrations and mode 4, 5 and 6 are stretching vibrations.
4.mode1, 3, 4 and 6 are highly symmetric.
5.mode1 is the umbrella vibration mode
6.Three bands would be expected in the experimental spectrum.
Atomic charges
charge on N atom =-1.125e
charge on H atom = 0.375e
N2
Calculation Method RB3LYP
Basis set 6-31G(d.p)
Final energy E(RB3LYP) -109.52359111 a.u.
Point group D*H
RMS gradient norm 0.02473091 a.u.
N-N bond length 1.10550 Å
N-N bond angle 180°
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401020D-13
Optimization completed.
-- Stationary point found.
N2 |
charge 0e
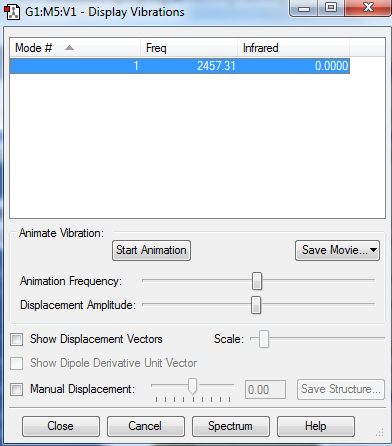
1. N2 is linear, 1 mode from 3N-5 is expected
H2
Calculation Method RB3LYP
Basis set 6-31G(d.p)
Final energy E(RB3LYP) -1.15928020 a.u.
Point group D*H
RMS gradient norm 0.09719500 a.u.
H-H bond length 0.60000 Å
H-H bond angle 180°
charge 0e
H2 |
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
Reactivity
E(NH3)= -56.44397188 a.u
2*E(NH3)= -112.8879438 a.u.
E(N2)= -109.52359111 a.u.
E(H2)= -1.15928020 a.u.
3*E(H2)= -3.4778406 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=0.11348791 a.u. = 297.9625077 kJ/mol
The gaseous reactants are more stable.
Cl2
Calculation Method RB3LYP
Basis set 6-31G(d.p)
Final energy E(RB3LYP) -920.34853293 a.u.
Point group D*H
RMS gradient norm 0.01404387 a.u.
Cl-Cl bond length 2.04174 Å
Cl-Cl bond angle 180°
charge 0e
Item Value Threshold Converged?
Maximum Force 0.000043 0.000450 YES
RMS Force 0.000043 0.000300 YES
Maximum Displacement 0.000121 0.001800 YES
RMS Displacement 0.000172 0.001200 YES
Predicted change in Energy=-5.277217D-09
Optimization completed.
-- Stationary point found.
Cl2 |
charge 0e
CO
Calculation Method RB3LYP
Basis set 6-31G(d.p)
Final energy E(RB3LYP) -113.28443706 a.u.
Point group C*V
RMS gradient norm 0.11032915 a.u.
C=O bond length 2.04174 Å
C=O bond angle 180°
Dipole moment 0.3395 debye
Charge atoms O -0.537e C 0.537e
Item Value Threshold Converged?
Maximum Force 0.000032 0.000450 YES
RMS Force 0.000032 0.000300 YES
Maximum Displacement 0.000012 0.001800 YES
RMS Displacement 0.000018 0.001200 YES
Predicted change in Energy=-3.956716D-10
Optimization completed.
-- Stationary point found.
CO |
Molecular Orbital
This is the 3σ orbital of CO, and also the HOMO of CO, which is made up of the 2p orbital of C and 2p orbital of O in phase, therefore it's a bonding orbital.
Because the 3σ mix with the 2σ orbital, 3σ has a higher energy than 1π orbital.
This is the pi* orbital of CO, and also the LUMO of CO, which is made up of the 2p orbital of C and 2p orbital of O out of phase, therefore it's an antibonding orbital
These two orbitals are 1 pi orbital, which are made of 2 2p orbitals of C and O in phase (bonding orbital), and they are degenerate in energy.