Rep:Mod:YHmodule2comp
The basic techniques of molecular mechanics and semi-empirical molecular orbital methods for structural and spectroscopic evaluations
BH3
| Calculation method | B3LYP |
| Basis set | LANL2MB |
| B-H distance/Å | 1.86592 ± 0.01 |
| H-B-H bond angle/° | 120 ± 0.1 |
| Point group | D3H |
MO analysis of BH3

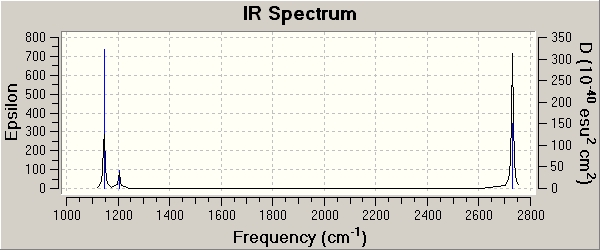
IR of BH3
BCl3
| Calculation method | B3LYP |
| Basis set | LANL2MB |
| B-Cl distance/Å | 1.87 |
| Cl-B-Cl bond angle/° | 120 ± 0.1 |
| Point group | D3H |
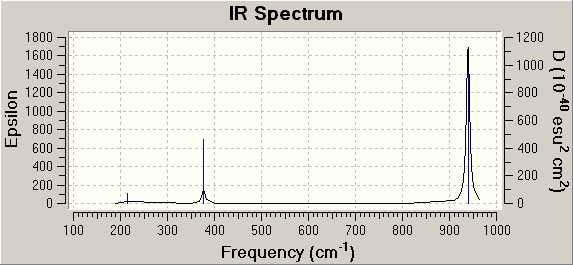
In carrying any computational analysis, the molecule to be studied is always optimsed first. This determines the optimium position of the nuclei for a given electronic configuration. A loose optimisation using a low accuracy basis e.g. 3-21G set will speed up the subsequent optimisation using the medium level basis set LANL2MB. After optimisation, a frequency analysis is carried out to confirm that the optimisation has found the minima structure. The frequency analysis is the second derivative of the potential energy surface, and when the second derivative is positive, a minumum has been achieved. The test is to see if there are any negative frequencies, since this implies that the optimisation calculation failed to find a critical point so it did not complete. In my case, all the frequencies were positive, confirming that the optimisation had worked.
The same method and basis set has to be used for each calculation to garner meaningful results. Basis sets determine the number of functions used to describe the electronic structure and should be consistent for the study of each molecule.
The optimised B-Cl bond length calculated by Gaussview was given as 1.86592Å but it is only accurate to ~0.01 Å. Therefore the bond length for comparison is 1.87Å. The literature bond length is 1.75Å. The optimised Cl-B-Cl bond angle was 120°. This is identical to the literature bond angle.[1]
For certain structures the optimised structure in Gaussview does not contain all the bonds expected. This is because Gaussview has an internal list of bond distances and if the computed bonds are outside of this Gaussview does not draw them in. This is most evident for inorganic bonds since they are typically longer than those found in organic complexes and are longer than the distance criteria set by Gaussview. This does not mean that the bonds don't exist since they are not just a line. A bond is an interaction between electrons...
The symmetry group of the ground state BCl3 structure is D3h and this is the same as what Gaussview used. Before the optimisation was carried out, the point group of the molecule was constrained to D3h and the tolerance was set to very high. This is to obtain accurate optimisation and frequency analysis because the atoms would be more fixed in space.
The optimisation calculation took 9seconds and the frequency analysis took 18 seconds when run on the laptop. There is a trade off between these short calculation times and the level of the basis set. For this relatively simple molecule it is acceptable to have a medium level basis set but this is not the case in the next section.
| No | Form of the vibration | Frequency/cm-1 | Intensity | symmetry of D2h point group |
|---|---|---|---|---|
| 1 |  |
214.13 | ||
| 2 |  |
214.13 | ||
| 3 |  |
376.94 | A2" | |
| 4 |  |
417.38 | A1' | |
| 5 |  |
939.47 | ||
| 6 |  |
937.47 |
IR of BCl3
Isomer of Mo(CO)4(L)2
In the second year inorganic lab course the cis and trans isomer of Mo(CO)4(L)2 where L=PPh3 were prepared and the IR spectra were analysed. For the computational analysis, the bulky PPh3 ligand is going to be replaced with PCl3. This is to reduce the computing time and make the calculation less expensive to run. Cl is chosen to replace Ph because it has a similar electronic contribution to bonding as pheyl groups and is still sterically quite large.
The number of CO vibrational bands active is related to the symmetry of the complex, four carbonyl absorption bands are expected from the compound with cis ligands and only one band is expected from the compound with trans ligands. This was seen in the experimental IR spectra and will be analysed in the computational IR spectra as well. Additionally, the structure and stability of the cis and trans isomer will be discussed.
The first step was to optimise the structures and then frequency analysis was undertaken.
Cis Isomer
| Calculation method | B3LYP |
| Basis set | LANL2DZ |
| Mo-C bond length/Å | 1.68 |
| Mo-P bond length/Å | 1.68 |
| P-Cl bond length/Å | 2.21 |
| Cl-Mo-Cl bond angle/° | 120 ± 0.1 |
| Point group | C1 |
| Bond | Computed/Å | Literature/Å |
| Mo-C | 1.68 | 1.58 |
| Mo-P | 1.68 | 1.58 |
| C=O | 1.68 | 1.58 |
| P-Cl | 2.21 | 1.97 |
| Bond | Computed/° | Literature/° |
| Cl-P-Cl | 100.8 | 102.0 |
| Mo | 117.0 | 118.4 |
| Mo | 123.0 | 121.4 |
| vibration | frequency/cm-1 | intensity |
|---|---|---|
 |
1945 | |
 |
1948 | |
 |
1958 | |
 |
2023 |
Trans Isomer
| Calculation method | B3LYP |
| Basis set | LANL2DZ |
| Mo-C bond length/Å | 1.86592 ± 0.01 |
| Mo-Cl bond length/Å | |
| Cl-Mo-Cl bond angle/° | 120 ± 0.1 |
| Point group | C1 |
| vibration | frequency/cm-1 | intensity |
|---|---|---|
 |
1950.29 | |
 |
1950.93 | |
 |
1977.21 | 0.64 |
 |
2030.99 | 3.78 |
Only one peak in the IR spectra is expected, but there are four frequencies shown in the table. This is due to the fact that Gaussview did not consider the correct point group of the molecule, . It considered the point group to be C1 so therefore all the CO ligands were not considered equivalent. However, the two vibrations at 1977 and 2030cm-1 are of very low intensities and are not visible in the IR spectrum. The other two vibrations occur at near equivalent wavenumber, Gaussview has considered the axial and equatorial CO stretches to be inequivalent which is incorrect.
Discussion of cis and trans isomer
The energy of the cis isomer was calculated to be -623.57707195 a.u. the energy of the trans isomer as -623.57603109 a.u., giving a small energy difference of -2.7kJ/mol. This implies that the cis isomer is more stable. However, the energies calculated by Gaussview have an error of ~10kJ/mol and once this is taken into account, the greater stability of one isomer over the other cannot be inferred. The cis isomer might be the more stable because two of the CO ligands are 90° apart from one another. Bonding in metal carbonyl complexes arise from donation of electron density from the metal filled d orbitals to the empty anti-bonding CO orbital. Since the CO are 90°, they do not compete with eachother for backbonding from the same MO d orbitals.
On the other hand, I predict that as the size of L ligand increases, the trans isomer will be more stable. In the case of Mo(CO)4L2 where L=PPh3, steric effects between the PPh3 ligands would destabilise the cis isomer.
Mini Project based on Ferrocene
For the project, I have chosen to study ferrocene. It consists of two cyclopentadienyl rings bound on opposite sides of the central Fe atom. The barrier for rotation of the Cp rings about the central Fe is very low so at room temperature, the rings are constantly rotating, between the staggered and the eclipsed form. I will start off with analysing the staggered and eclipsed form of ferrocene to study their relative stability. From this point, I aim to study the effect of changing the cyclopentadiene ligang by replacing the H with electron donating and electron withdrawing groups and analyse the natural bond order of the molecules.
Staggered and Eclipsed Ferrocene
I will optimise the two structures of ferrocene and then carry out frequency analysis. Since there is a Fe atom to consider, I will use the high level basis set LANL2DZ and pseudo-potentials because the relativistic effects of Fe cannot be recovered by using the Schrodinger equation. Staggered and eclipsed. Staggered is a Transition State, shown by freq analysis. One negative frequency.
References and citations
- ↑ S. Gierszal, J. Galica and E. Mis-Kuzmin ska, Physica Scripta, 2003, 67.






