Rep:Mod:XYZ12348
Bonding and Molecular Orbitals in Main Group Compounds
Introduction
Gaussian modelling can be used in order to optimize a molecule and thereby collect thermodynamic information and kinetic data
Optimisation of BH3
Gaussian modelling can be used in order to optimize a molecule and thereby collect thermodynamic information and kinetic data. The molecule which has been investigated is BH3 and an image of the non-optimised form is shown below.
During the optimisation of a molecule, if we assume that the B and H nuclei have a given position, then we can solve the Schrodinger equation and therefore determining the electron density and energy of the nuclei in that position, known as the SCF part of the Schrodinger equation. From this, we can then determine the equation for the particular position of the nuclei and this can be achieved by moving the nuclei between different SCF's until the lowest energy geometry is found which is known as the optimised geometry. The first optimisation carried out used a B3LYP method and the optimisation data for BH3 can be found here:File:BH3 OPT.LOG. An image for the optimised molecule is shown below.
The B-H bond distance for this molecule is: 1.19349Å
The H-B-H bond angle for this molecule is: 120
The table below summaries the information gained from the optimisation of the molecule. One piece of information shown in the table is the basis set and this shows the level of accuracy of the calculation. This particular basis set, 3-21G, has a low accuracy, although this does mean that the speed of the calculations are fast, in this particular case 10 seconds.
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3 LYP |
| Basis Set | 3-21G |
| Final Energy (a.u) | -26.46226338 |
| Gradient (a.u) | 0.00020672 |
| Dipole moment(D) | 0.0000 |
| Point group | D3H |
| Length of Calculation (s) | 10 |
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Optimisation plots of BH3
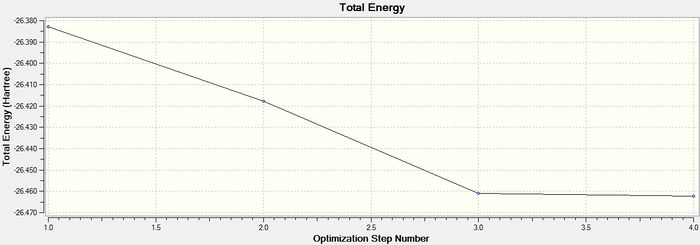
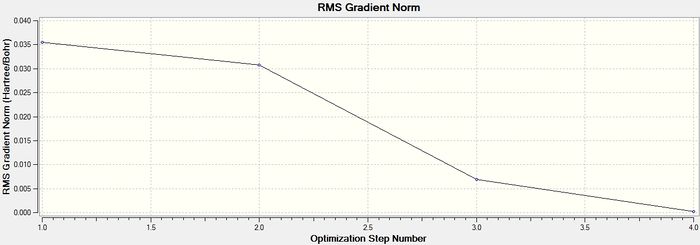
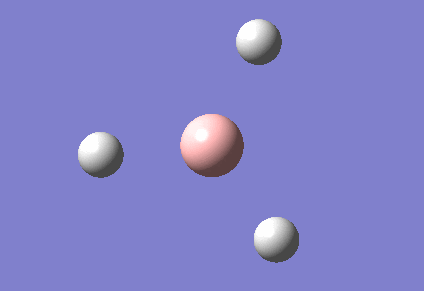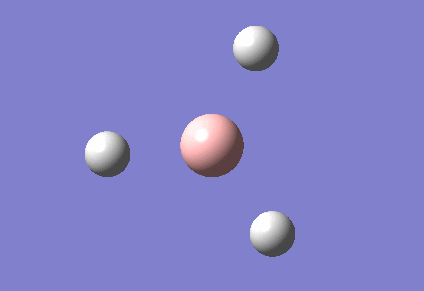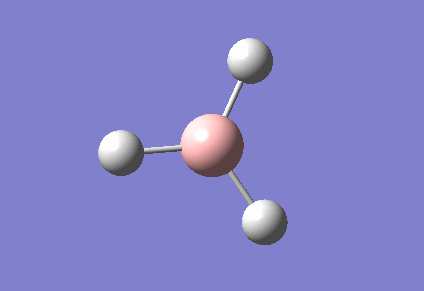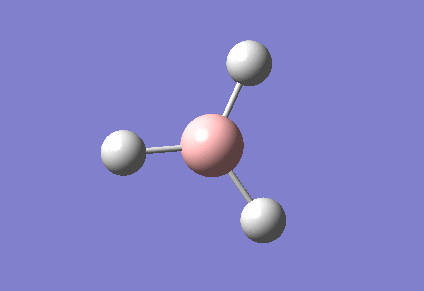
A better Gaussian model of BH3
A 6-13G optimisation of BH3 is shown here:File:BH3 OPT 6 13GJND.LOG
The B-H bond length is: 1.19143A
The H-B-H bond angle is:120
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3 LYP |
| Basis Set | 6-13G(d,p) |
| Final Energy (a.u) | -26.61532252 |
| Gradient (a.u) | 0.00021672 |
| Dipole moment(D) | 0.0000 |
| Point group | D3H |
| Length of Calculation (s) | 7 |
Optimisation of TIBr3
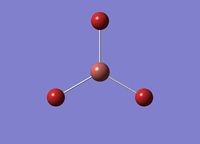

The link for the optimised TIBr3 molecule can be found here: File:Log 69180 TIbr3optimisation.log
The TI-Br bond length is: 2.65095
The Br-TI-Br angle is: 120
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3 LYP |
| Basis Set | LanL2DZ |
| Final Energy (a.u) | -91.21812851 |
| Gradient (a.u) | 0.00000090 |
| Dipole moment(D) | 0.0000 |
| Point group | D3H |
| Length of Calculation (s) | 23.5 |
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Optimisation of BBr3
The link for the optimised BBr3 molecule can be found here: File:BBr3 optimistion JND.log
The B-Br bond length is: 1.93396
The Br-B-Br angle is:120
| File type | .log |
| Calculation type | FOPT |
| Calculation method | RB3 LYP |
| Basis Set | Gen |
| Final Energy (a.u) | -64.43645296 |
| Gradient (a.u) | 0.00000382 |
| Dipole moment(D) | 0.0000 |
| Point group | D3H |
| Length of Calculation (s) | 25.6 |
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.026780D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
