Rep:Mod:XYZ12345
Bonding and Molecular Orbitals in Main Group Compounds
Optimisation of BH3
The BH3 molecule (shown below) was optimised [OPT] using B3LYP as method and a basis set of 3-21G.
The optimisation file is linked to File:BH3 OPT.LOG
Optimised B-H bond distance for the BH3 molecule: 1.19359 Å
Optimised H-B-H bond angle for the BH3 molecule: 120o
| File type | bh3_opt |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis set | 3-21G |
| Final energy in atomic units (au) | -26.46226338 |
| Gradient | 0.00020672 |
| Dipole moment (Debeye) | 0 |
| Point group of the molecule | D3H |
| Duration of calculation (s) | 10.0 |
The table below shows both the convergence of the force and displacements after the first optimisation:
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
The process of optimisation is to solve the Shrodinger equation for the electrons and differing nuclear posisitons, until the energy obtained is at a minimum. The molecule will experience forces F(R) when a change in position detla R causes a change in energy E(R). When this criteria is not fulfilled i.e the nuclear and electron are in equilibrium, the molecule will experience no forces. The force is related to the potential energy and nuclei displacement through F= -dE/dr [where E is the potential energy and r is the nuclei displcacement]. Thus, at equilibrium structure the gradient of the curve, which is the first derivative dE/dr would be zero. Hence, the force experienced at equilibrium is zero.
As seen in the table above, the maximum force and displacement values are lower than the threshold value. This reflects the molecules movement along the PES surface, towards the equilibrium position and hence experiencing a lower force.
Energy and gradient changes during optimisation of BH3 molecule
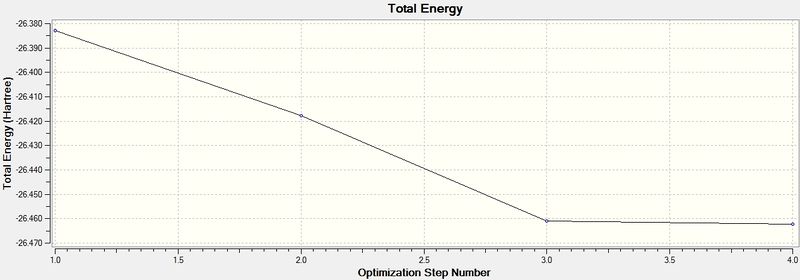
The graph above represents the program traversing Potential Energy Surface (PES) to attain the minimum energy structure of BH3.
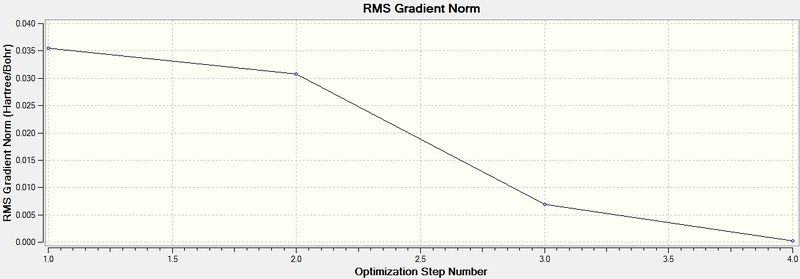
The graph above represents the Root Mean Square Gradient, showing the clearly the gradient tending to zero as the equilibrium structure of BH3 is obtained.
Depiction of structure at each optimisation step



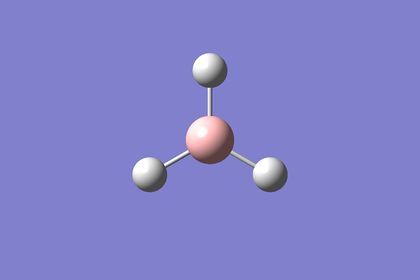
As can be seen from the first image no bonds in the BH3 structure are shown. A bond can be defined as the attractive force which holds together two or more atomic species.
Higher level basis set
The link for the optimised 6-31G(d,p) BH3 is file is File:BH3 OPT631G.LOG

Optimised B-H bond distance for the BH3 molecule: 1.19143 Å
Optimised H-B-H bond angle for the BH3 molecule: 120o
| File type | BH3_OPT631G |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy in atomic units (au) | -26.61532252 |
| Gradient | 0.00021672 |
| Dipole moment (Debeye) | 0 |
| Point group of the molecule | D3H |
| Duration of calculation (s) | 9.0 |
The table below shows both the convergence of the force and displacements after the 6-31G (d,p) optimisation:
Item Value Threshold Converged?
Maximum Force 0.000433 0.000450 YES
RMS Force 0.000284 0.000300 YES
Maximum Displacement 0.001702 0.001800 YES
RMS Displacement 0.001114 0.001200 YES
Predicted change in Energy=-1.189019D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1914 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1914 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1914 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
TlBr3 optimisation

The link for the optimised file is File:TlBr3LanL2DZ OPT.log
Optimised Tl-Br bond distance for the TlBr3 molecule: 2.65095 Å
Optimised Br-Tl-Br bond angle for the TlBr3 molecule: 120o
| File type | log_69179 |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis set | LANL2DZ |
| Final energy in atomic units (au) | -91.21812851 |
| Gradient | 0.00000090 |
| Dipole moment (Debeye) | 0 |
| Point group of the molecule | D3H |
| Duration of calculation (s) | 21.8 |
The table below shows both the convergence of the force and displacements after the LanL2DZ optimisation:
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084075D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
BBr3 optimisation

The link for the optimised file is File:BBr3GEN OPToc.log
Optimised B-Br bond distance for the BBr3 molecule: 1.93396 Å
Optimised Br-B-Br bond angle for the BBr3 molecule: 120o
| File type | BBr3GEN_OPToc |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis set | Gen |
| Final energy in atomic units (au) | -64.43645296 |
| Gradient | 0.00000382 |
| Dipole moment (Debeye) | 0 |
| Point group of the molecule | D3H |
| Duration of calculation (s) | 21.4 |
The table below shows both the convergence of the force and displacements after the Gen optimisation:
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027672D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
