Rep:Mod:Wiki01386810
NH3 optimization
Summary Details
calculation method: RB3LYP
basis set: 6-31G(d,p)
final energy: -56.55776873 au
RMS gradient: 0.00000485 au
point group: C3V
optimized N-H bond length: 1.01798 A
optimized bong angle: 105.74115
The charge on the Nitrogen atom is -1.125 and the charge on the hydrogen atom is 0.375. As the nitrogen atom is much more electronegative than the hydrogen atoms it pulls electron density towards itself therefore you'd expect it to have a negative charge while the hydrogen's would be electron deficient and so positively charged
Item Table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
Link to data:media:NH3_OPT_01386810.LOG
NH3 Dynamic Image
NH |
Link to file:(File:NH3 OPT 01386810.LOG)
NH3 Vibration Data
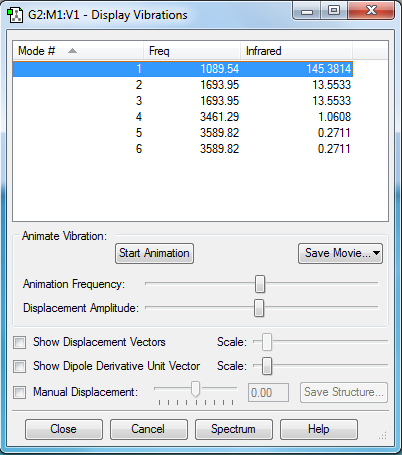 no negative frequencies shown therefore the optimization worked
no negative frequencies shown therefore the optimization worked
Analysis Questions&Answers
how many modes do you expect from the 3N-6 rule?
3(4)-6= 6 therefore you'd expect 6 modes
which modes are degenerate?
modes 2 and 3 have the same energy and modes 5 and 6 have the same energy
which modes are "bending" vibrations and which are "bond stretch" vibrations?
1,2 and 3 are bending while 4,5 and 6 are stretching
which mode is highly symmetric?
mode 4
one mode is known as the "umbrella" mode, which one is this?
mode 1
how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2 bands would be expected in an experimental spectrum of ammonia which would be for modes 1 & 2/3. 2 & 3 have the same energy so have the same band. The rest of the modes are not IR active as there is no net change in dipole moment during vibrations
N2 optimization
Summary Details
calculation method: RB3LYP
basis set: 6-31G(d,p)
final energy: -109.52412868 au
RMS gradient: 0.00000060 au
Point Group: D∞h
optimized N-N bond length: 1.10550 A optimized bong angle: 180
Item Table
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
Link to data:media:N2 OPT.LOG
N2 Dynamic Image
N |
Link to file:(File:N2 OPT.LOG)
N2 Vibration Data
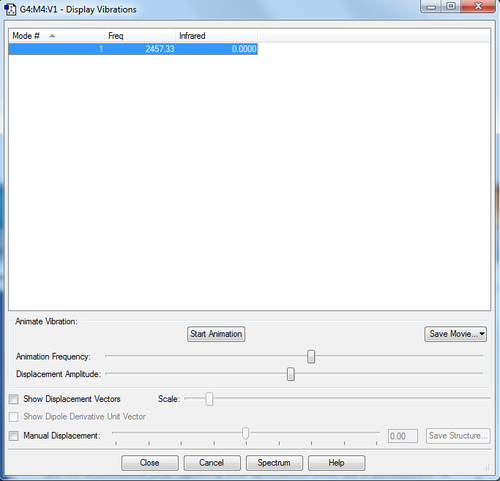 no negative frequencies shown therefore the optimization worked. For this molecule there is no change in dipole moment as electronegativites of atom within the molecule are the same and so intensity for peak is very small and the peak would be seen on a spectrum
no negative frequencies shown therefore the optimization worked. For this molecule there is no change in dipole moment as electronegativites of atom within the molecule are the same and so intensity for peak is very small and the peak would be seen on a spectrum
H2 optimization
Summary Details
calculation method: RB3LYP
basis set: 6-31G(d,p)
final energy: -1.17853936 au
RMS gradient: 0.00000017 au
point group: D∞h
optimized H-H bond length: 0.74279 A optimized bong angle: 180
Item Table
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
Link to data:media:H2 OPT.LOG
H2 Dynamic Image
H |
Link to file:(File:H2 OPT.LOG)
H2 Vibration Data
 no negative frequencies shown therefore the optimization worked. For this molecule there is no change in dipole moment as electronegativites of atom within the molecule are the same and so intensity for peak is very small and the peak would be seen on a spectrum
no negative frequencies shown therefore the optimization worked. For this molecule there is no change in dipole moment as electronegativites of atom within the molecule are the same and so intensity for peak is very small and the peak would be seen on a spectrum
Haber-Bosch Process Analysis
below is the energy values used to determine the energy for the reaction of N2 + 3H2 -> 2NH3
E(NH3)= -56.55776873 2*E(NH3)= -113.11553746 E(N2)= -109.52412868 E(H2)= -1.17853936 3*E(H2)= -3.53561808 ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579070= -146.48 kJ mol-1
The negative value for ΔE suggests that this reaction is exothermic therefore this means that the product is lower in energy than the products so ammonia is more stable than the reactants
S2 optimization
Summary Details
calculation method: RB3LYP
basis set: 6-31G(d,p)
final energy: -796.32599779 au
RMS gradient: 0.00000372 au
point group: D∞h
optimized S-S bond length: 1.92943 A
optimized bong angle: 180
Charges on both S atoms are 0 as they are the same atom and have the same electronegativity values so both pull electron density towards each other the same amount
Item Table
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000006 0.000300 YES Maximum Displacement 0.000011 0.001800 YES RMS Displacement 0.000016 0.001200 YES
Link to data:media:S2 OPT 2.LOG
S2 Dynamic Image
S |
Link to file:(File:S2 OPT 2.LOG)
S2 Vibration Data
 no negative frequencies shown therefore the optimization worked. For this molecule there is no change in dipole moment as electronegativites of atom within the molecule are the same and so intensity for peak is very small and the peak would be seen on a spectrum
no negative frequencies shown therefore the optimization worked. For this molecule there is no change in dipole moment as electronegativites of atom within the molecule are the same and so intensity for peak is very small and the peak would be seen on a spectrum
Molecular Orbital Analysis
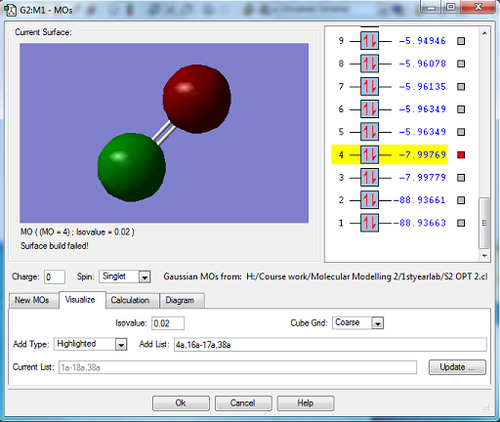
These two images above show both the bonding (2σg) and anti-bonding (2σu*) molecular orbitals formed when the 2s atomic orbitals on both sulfur atoms combine. As shown in the image these orbitals are low in energy relative to the orbitals and because they are so low in energy they dont contribute much to chemical bonding. The bonding orbital is lower in energy than the anti-bonding orbital (0.0001au difference between them) and both molecular orbitals are fully occupied by electrons . Also shown in the image is the energies for the 1s molecular orbitals which i much lower in energy than the 2s orbitals and are also not involved in chemical bonding.
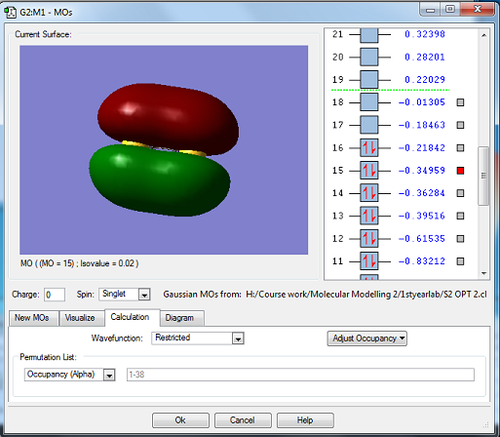
These two images above show both the HOMO (highest occupied molecular orbital) and LUMO (lowest unoccupied molecular orbital, meaning its unfilled) which are formed through the combination of the 3p atomic orbitals on both sulfur atoms and have the label 3πg* (two p orbitals on one atom combine with another two from the other atom orthogonal to the inter nuclear axis in the formation of these MOs). These molecular orbitals are involved in chemical bonding and are very similar in energy. The label of these orbitals shown depends on which p-orbitals were involved in there formation. Both the HOMO and LUMO are anti-bonding orbitals and because the HOMO is an anti-bonding interaction this makes the corresponding bond very reactive
This image shows the molecular orbital 3πu meaning that this is the bonding orbital (meaning that the 3p orbitals which combined to form this were in phase) which corresponds to the HOMO and LUMO as for . This molecular orbital is relatively high in energy and is filled with electrons