Rep:Mod:WMX1205
Week 1: Compulsory Section BH3
BH3 optimization
The optimization of BH3 was carried out by the GaussView calculation with the following settings:
Molecule
BH molecule |
Summary
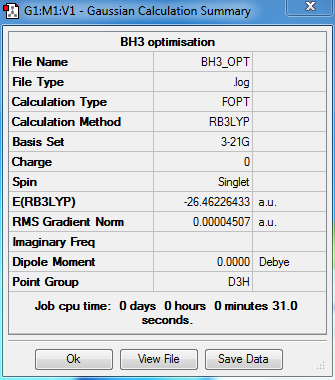
Output information
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000059 0.000300 YES
Maximum Displacement 0.000352 0.001800 YES
RMS Displacement 0.000230 0.001200 YES
Predicted change in Energy=-4.580970D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1945 -DE/DX = -0.0001 !
! R2 R(1,3) 1.1945 -DE/DX = -0.0001 !
! R3 R(1,4) 1.1945 -DE/DX = -0.0001 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Bond information
| Bond Distance | 1.19453 Å |
| Bond Angle | 120.000 |
File link
BH3 optimization with 6-31G(d,p) basis set
The optimization with better basis set of BH3 was carried out by the GaussView calculation with the following settings:
Molecule
BH3 with 6-31G(d,p) basis set |
Summary
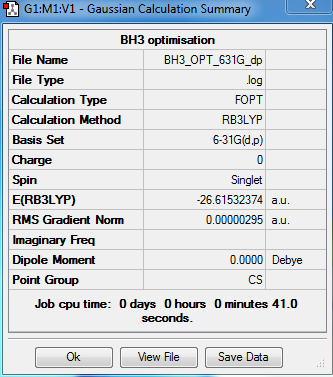
Output information
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000024 0.001800 YES
RMS Displacement 0.000015 0.001200 YES
Predicted change in Energy=-1.980661D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0002 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0002 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9997 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Bond information
| Bond Distance | 1.19232 Å |
| Bond Angle | 120.000 |
File link
File:WENMIN BH3 OPT 631G DP.LOG
Total energy of BH3
Total energy of 3-21G optimized BH3: E = -26.46226433 a.u.
Total energy of 6-31G optimized BH3: E = -26.61532374 a.u.
TlBr3 optimization
The optimization with restricted symmetry of TlBr3 was carried out by the HPC with the following settings:
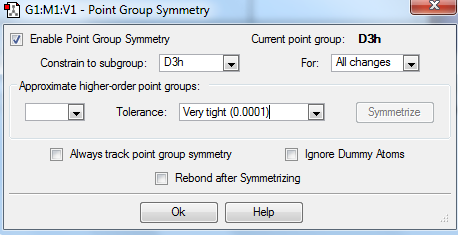
Molecule
TlBr molecule |
Summary
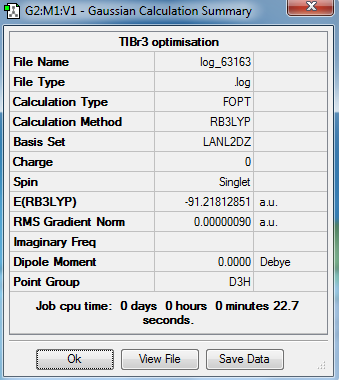
Output information
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.083028D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Bond information
| Bond Distance | 2.65095 Å |
| Bond Angle | 120.000 |
Compared with the literature value[1], Tl-Br distance of 2.512 Å, the bond distance optimized by the GaussView is a little larger.
File link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20479
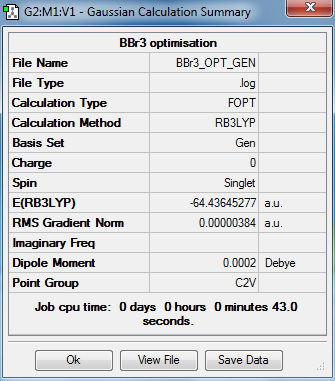
BBr3 optimization
The optimization of BBr3 was carried out by the GaussView calculation with the following settings:
Molecule
BBr molecule |
Summary
Output information
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000024 0.001200 YES
Predicted change in Energy=-4.098023D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.9339 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0022 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9956 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0022 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Bond information
| Bond Distance | 1.93394 Å |
| Bond Angle | 120.000 |
File link
Comparison of BH3, BBr3 and TlBr3 optimization
| BH3 | 1.19232 Å |
| BBr3 | 1.93394 Å |
| TlBr3 | 2.65095 Å |
It is shown that the bond length increases down the table. The equilibrium bond length in a molecule is the distance between the centers of the two bonded atoms.[2] To a reasonable approximation, the bond length can be partitioned into the radius contributions from each atom. From BH3 ligand to BBr3 ligand, the size of Br atom is larger than that of H atom, leading to a longer bond. Similarly, changing central element from BBr3 to TlBr3, the bond length increases due to larger size of Tl atom (the atom size increases down the group e.g. B<Al<Ga<In<Tl).
Both H and B atoms are small. However, H atom has only one (valence) electron while B atom has five electrons (three valence electrons). The fist ionization energy of H atom is larger than that of B atom (H: 1312 kJ/mol; B: 801 kJ/mol). [3] This is because the valence electron of B atom is further away from the nucleus and experiences a relatively weaker Coulombic attraction, when compared with H atom.
B atom and Tl atom belong to the same group - Group 13. They both have one valence electron, but they differ in many ways. Boron is nonmetallic while thallium is metallic. B has higher first ionization energy than Tl because B is small atom with electrons being closer to the nucleus and experiencing stronger Coulombic attracting. Generally, the ionization energy decreases down the group. Boron has a very high melting point.
Sometimes GaussView doesn't display bonds where we expect, it doesn't necessarily mean that there's no bond. GaussView draws bonds based on a distance critera. When there's no bond, it could be due to that the bond distance exceeds a pre-defined value. Bond is attraction among atoms or molecules. Atoms or molecules either share or donate electrons, or in other means, to achieve most stable configuration (e.g. Octet rule, 18-electron rule) while forming the bond.
BH3 frequency
The frequency calculation of BH3 molecule was carried out by the GaussView with the following settings:
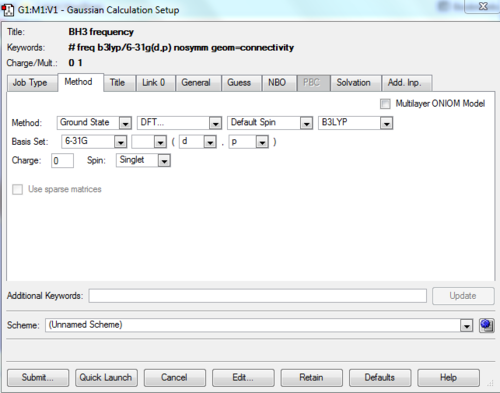
Summary
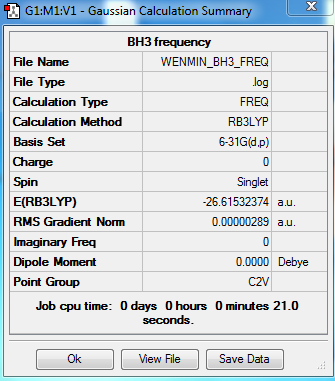
Output information
Low frequencies --- -19.0010 0.0007 0.0009 0.0009 12.0258 12.0968
Low frequencies --- 1162.9746 1213.1729 1213.2338
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 1162.9746 1213.1729 1213.2338
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9986 0.9601 0.9602
IR Inten -- 92.5518 14.0571 14.0606
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.16 0.00 0.10 0.00 -0.10 0.00 0.00
2 1 0.00 0.00 -0.57 0.00 0.08 0.00 0.81 0.00 0.00
3 1 0.00 0.00 -0.57 0.39 -0.59 0.00 0.14 -0.39 0.00
4 1 0.00 0.00 -0.57 -0.39 -0.59 0.00 0.14 0.39 0.00
4 5 6
A A A
Frequencies -- 2582.2780 2715.4300 2715.4555
Red. masses -- 1.0078 1.1273 1.1273
Frc consts -- 3.9595 4.8976 4.8977
IR Inten -- 0.0000 126.3279 126.3232
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.00 0.11 0.00 0.00 0.00 0.11 0.00
2 1 0.00 -0.58 0.00 0.02 0.00 0.00 0.00 -0.81 0.00
3 1 0.50 0.29 0.00 -0.60 -0.36 0.00 -0.36 -0.19 0.00
4 1 -0.50 0.29 0.00 -0.60 0.36 0.00 0.36 -0.19 0.00
Summary of motion, frequency and intensity of each vibration
IR spectrum
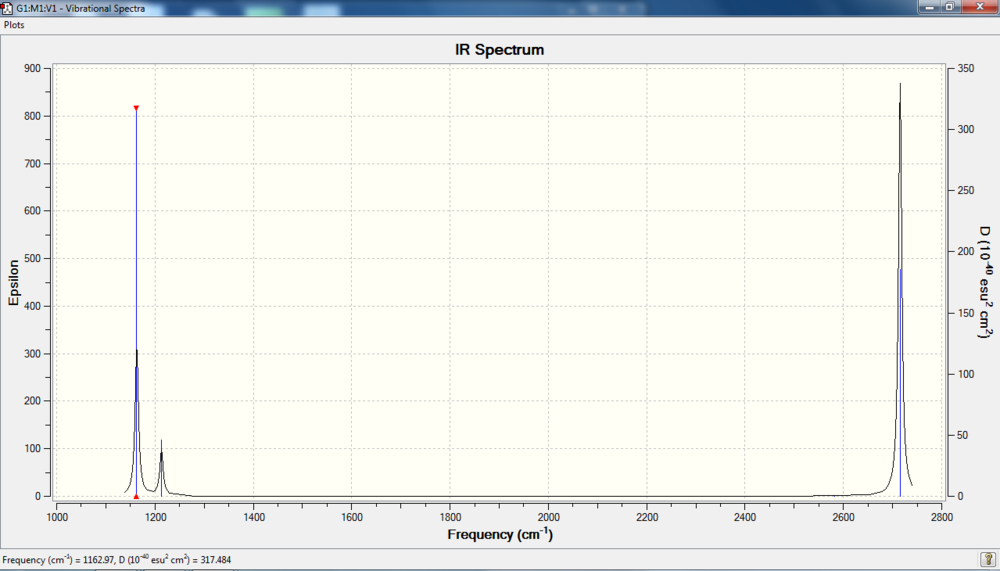
There are six vibrations but only three peaks. Look at the intensity. The intensity of vibration 3 is 0, which shows no peak on the IR spectrum. As for vibration 2&3 and 5&6, these two pairs have very similar peak intensity. They overlap on the spectrum, showing as "one peak" each.
File link
TlBr3 frequency
The frequency calculation of TlBr3 molecule was carried out by the HPC with the following settings:
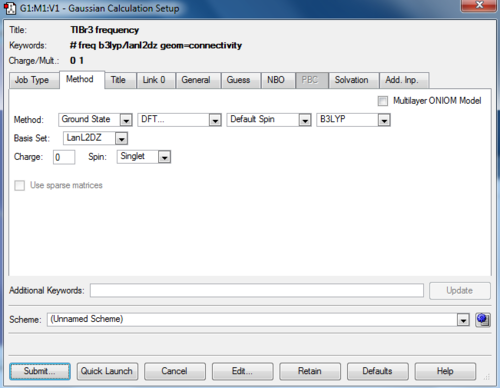
Summary
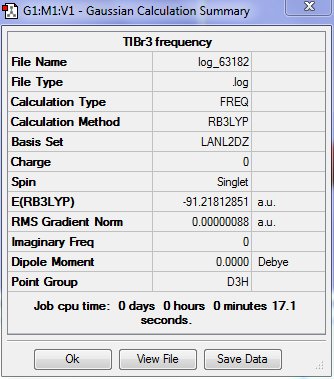
Output information
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367
Low frequencies --- 46.4289 46.4292 52.1449
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
E' E' A2"
Frequencies -- 46.4289 46.4292 52.1449
Red. masses -- 88.4613 88.4613 117.7209
Frc consts -- 0.1124 0.1124 0.1886
IR Inten -- 3.6867 3.6867 5.8466
Atom AN X Y Z X Y Z X Y Z
1 81 0.00 0.28 0.00 -0.28 0.00 0.00 0.00 0.00 0.55
2 35 0.00 0.26 0.00 0.74 0.00 0.00 0.00 0.00 -0.48
3 35 0.43 -0.49 0.00 -0.01 -0.43 0.00 0.00 0.00 -0.48
4 35 -0.43 -0.49 0.00 -0.01 0.43 0.00 0.00 0.00 -0.48
4 5 6
A1' E' E'
Frequencies -- 165.2685 210.6948 210.6948
Red. masses -- 78.9183 101.4032 101.4032
Frc consts -- 1.2700 2.6522 2.6522
IR Inten -- 0.0000 25.4830 25.4797
Atom AN X Y Z X Y Z X Y Z
1 81 0.00 0.00 0.00 0.42 0.00 0.00 0.00 0.42 0.00
2 35 0.00 -0.58 0.00 0.01 0.00 0.00 0.00 -0.74 0.00
3 35 0.50 0.29 0.00 -0.55 -0.32 0.00 -0.32 -0.18 0.00
4 35 -0.50 0.29 0.00 -0.55 0.32 0.00 0.32 -0.18 0.00
Summary of motion, frequency and intensity of each vibration
IR spectrum
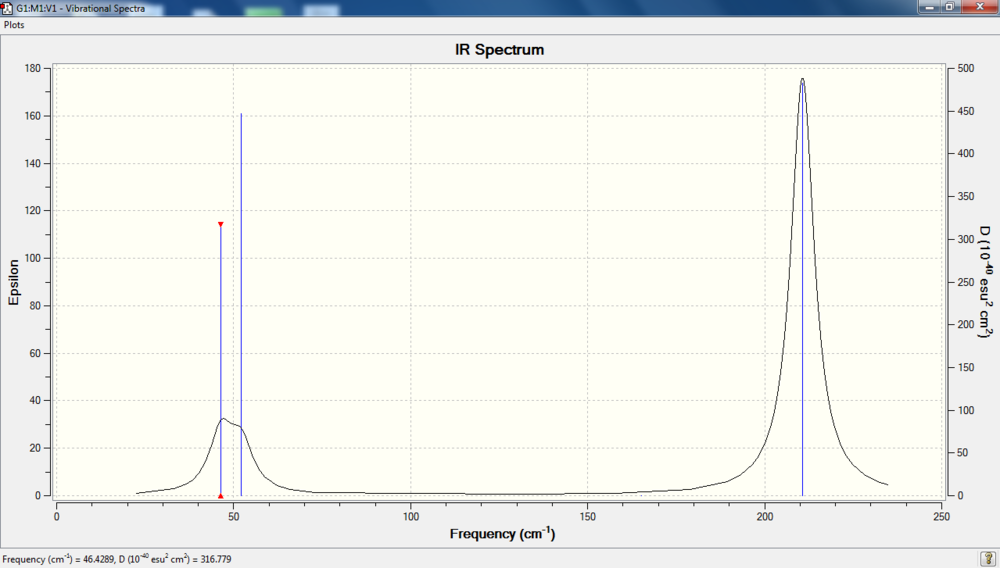
File link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20490
Comparison of BH3 and TlBr3 frequency
| No. | BH3 | TlBr3 |
|---|---|---|
| 1 | 1162.97 | 46.43 |
| 2 | 1213.17 | 46.43 |
| 3 | 1213.23 | 52.14 |
| 4 | 2582.28 | 165.27 |
| 5 | 2715.43 | 210.69 |
| 6 | 2715.46 | 210.69 |
The frequency of BH3 is lower than that of TlBr3. This is due to weaker bond of TlBr3. Both Tl and Br atoms are large so they are further away from the nucleus and experience weaker Coulomb attraction, resulting in a weaker bond. Hence TlBr3 has lower frequencies.
Both have six mode vibrations, but only three peaks with reasons stated above. There's a reordering of modes. BH3 has its lowest frequency at a'2 (mode 1) while TlBr3 has its lowest frequency at degenerate mode 1 and 2. Additionally, both of them have zero intensity for a'1 (mode 4).
Q&A
Why must you use the same method and basis set for both the optimization and frequency analysis calculations?
Under different method or basis set, the calculation from GaussView differs significantly. Take BH3 as an example, the energy under 3-21G basis set is -26.46226433 a.u. and the energy under 6-31G basis set is -26.61532374 a.u. The difference is 0.15305941 a.u., which is 401.86 kJ/mol. Comparing results under different settings is meaningless and worthless.
What is the purpose of carrying out a frequency analysis?
When we're doing the optimization calculation, we are doing the first derivative of the potential energy surface, which tells us the maximum (transition state) and minimum (ground state) with zero slope. However, we don't know which one is the maximum or minimum. That's when we need to take the second derivative to determine. When the second derivative is positive, we get a minimum and maximum if the second derivative is negative. What frequency analysis is mainly calculation of the second derivative. So if the frequencies are all positive, that indicates we have a minimum. If one of them is negative, we have a transition state. When there's more than one negative frequencies, it means unsuccessful finding of a critical point, indicating a failed optimization calculation. So frequency analysis checks the result of optimization. It also provide IR spectrum and Raman modes that are helpful.
What do the "Low frequencies" represent?
"Low frequencies" is an kind of indicator of the tightness of optimization. The low frequencies should be close to zero. If they are not close to zero, it may be a signal that we need to do a tighter optimization.
BH3 Molecular orbitals
MOs of BH3
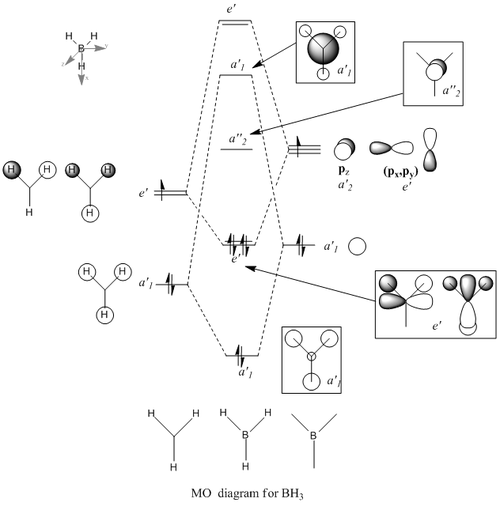
Real MOs
 |
 |
 |
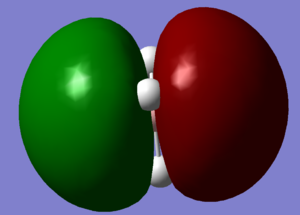 |
| a'1 | e' | e' | a'2' |
File link
DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20520
NH3 Molecular orbitals
The optimization and frequency calculation of TlBr3 molecule was carried out by the GaussView.
Molecule
NH molecule |
Summary
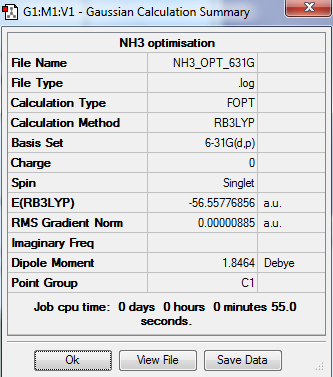
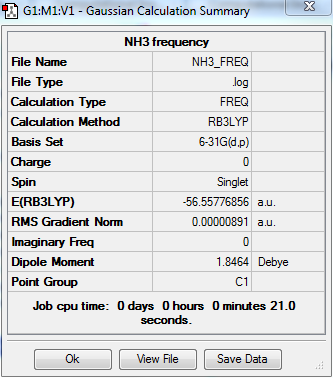
Output information
Optimization output information
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629731D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency output information
Low frequencies --- -30.7014 0.0010 0.0015 0.0022 20.2662 28.2997 Low frequencies --- 1089.5562 1694.1246 1694.1863
IR spectrum
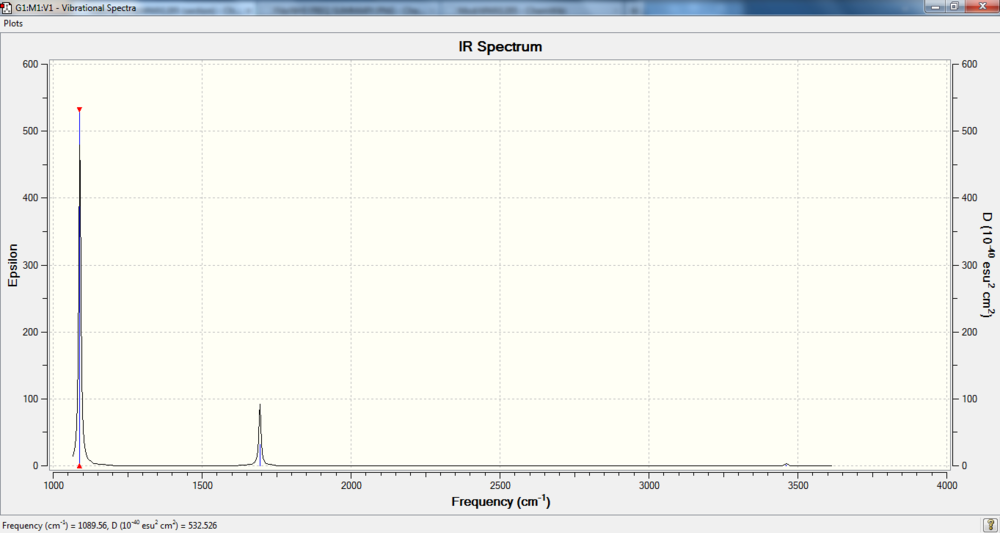
Charge distribution
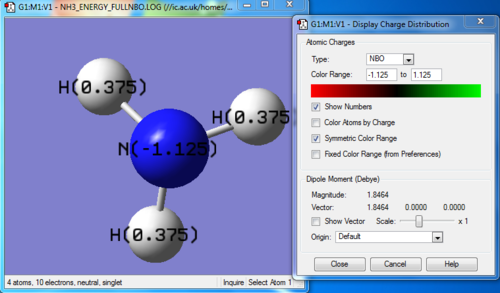
NBO analysis
Natural populations
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12515 1.99982 6.11104 0.01429 8.12515
H 2 0.37505 0.00000 0.62250 0.00246 0.62495
H 3 0.37505 0.00000 0.62250 0.00246 0.62495
H 4 0.37505 0.00000 0.62249 0.00246 0.62495
=======================================================================
* Total * 0.00000 1.99982 7.97852 0.02166 10.00000
Bond orbital
Occupancies Lewis Structure Low High
Occ. ------------------- ----------------- occ occ
Cycle Thresh. Lewis Non-Lewis CR BD 3C LP (L) (NL) Dev
=============================================================================
1(1) 1.90 9.99429 0.00571 1 3 0 1 0 0 0.00
-----------------------------------------------------------------------------
Structure accepted: No low occupancy Lewis orbitals
-------------------------------------------------------- Core 1.99982 ( 99.991% of 2) Valence Lewis 7.99447 ( 99.931% of 8) ================== ============================ Total Lewis 9.99429 ( 99.943% of 10) ----------------------------------------------------- Valence non-Lewis 0.00000 ( 0.000% of 10) Rydberg non-Lewis 0.00571 ( 0.057% of 10) ================== ============================ Total non-Lewis 0.00571 ( 0.057% of 10) --------------------------------------------------------
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
-0.0001 -0.4986 -0.0059 0.0000 -0.2910
0.0052 0.8155 0.0277 0.0000 0.0000
0.0281 0.0000 0.0000 0.0032 0.0082
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0000 0.0072 -0.0289 0.0000
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4077 0.0138 0.7062 0.0240
0.0140 0.0243 0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 -0.0250
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2909
-0.0052 0.4077 0.0138 -0.7062 -0.0239
0.0140 -0.0243 -0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 0.0250
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 -0.8618
0.0505 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0269 0.0155
6. (0.00000) RY*( 1) N 1 s( 99.98%)p 0.00( 0.02%)d 0.00( 0.00%)
7. (0.00000) RY*( 2) N 1 s(100.00%)
Second order perturbation theory analysis of Fock Matrix in NBO basis
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
within unit 1 5. LP ( 1) N 1 / 16. RY*( 1) H 2 1.01 1.43 0.034 5. LP ( 1) N 1 / 17. RY*( 2) H 2 0.67 2.17 0.034 5. LP ( 1) N 1 / 20. RY*( 1) H 3 1.01 1.43 0.034 5. LP ( 1) N 1 / 21. RY*( 2) H 3 0.67 2.17 0.034 5. LP ( 1) N 1 / 24. RY*( 1) H 4 1.01 1.43 0.034 5. LP ( 1) N 1 / 25. RY*( 2) H 4 0.67 2.17 0.034
Natural bond orbital
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99909 -0.60417
2. BD ( 1) N 1 - H 3 1.99909 -0.60417
3. BD ( 1) N 1 - H 4 1.99909 -0.60416
4. CR ( 1) N 1 1.99982 -14.16768
5. LP ( 1) N 1 1.99721 -0.31756 24(v),16(v),20(v),17(v)
21(v),25(v)
6. RY*( 1) N 1 0.00000 1.20799
7. RY*( 2) N 1 0.00000 3.73005
Bond information
| Bond Distance | 1.01797 Å |
| Bond Angle | 105.748 |
File link
Optimization:File:WENMIN NH3 OPT 631G.LOG
Frequency:File:WENMIN NH3 FREQ.LOG
NBO:File:WENMIN NH3 ENERGY FULLNBO.LOG
NH3BH3 analysis
The optimization and frequency calculation of NH3BH3 molecule was carried out by the GaussView.
Molecule
NHBH molecule |
Summary
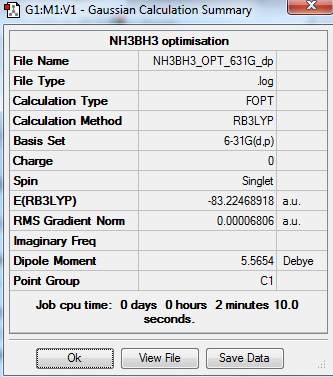
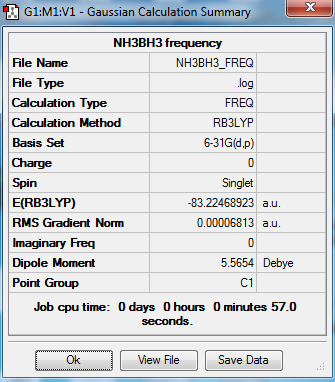
Output information
Optimization output information
Item Value Threshold Converged?
Maximum Force 0.000137 0.000450 YES
RMS Force 0.000063 0.000300 YES
Maximum Displacement 0.000606 0.001800 YES
RMS Displacement 0.000336 0.001200 YES
Predicted change in Energy=-1.994008D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.0186 -DE/DX = -0.0001 !
! R2 R(2,8) 1.0186 -DE/DX = -0.0001 !
! R3 R(3,8) 1.0186 -DE/DX = -0.0001 !
! R4 R(4,7) 1.2101 -DE/DX = -0.0001 !
! R5 R(5,7) 1.2101 -DE/DX = -0.0001 !
! R6 R(6,7) 1.2101 -DE/DX = -0.0001 !
! R7 R(7,8) 1.668 -DE/DX = -0.0001 !
! A1 A(4,7,5) 113.8693 -DE/DX = 0.0 !
! A2 A(4,7,6) 113.8721 -DE/DX = 0.0 !
! A3 A(4,7,8) 104.6003 -DE/DX = 0.0 !
! A4 A(5,7,6) 113.8747 -DE/DX = 0.0 !
! A5 A(5,7,8) 104.6003 -DE/DX = 0.0 !
! A6 A(6,7,8) 104.5984 -DE/DX = 0.0 !
! A7 A(1,8,2) 107.87 -DE/DX = 0.0 !
! A8 A(1,8,3) 107.8652 -DE/DX = 0.0 !
! A9 A(1,8,7) 111.0329 -DE/DX = 0.0 !
! A10 A(2,8,3) 107.8697 -DE/DX = 0.0 !
! A11 A(2,8,7) 111.0286 -DE/DX = 0.0 !
! A12 A(3,8,7) 111.0291 -DE/DX = 0.0 !
! D1 D(4,7,8,1) -179.9867 -DE/DX = 0.0 !
! D2 D(4,7,8,2) -59.9839 -DE/DX = 0.0 !
! D3 D(4,7,8,3) 60.0161 -DE/DX = 0.0 !
! D4 D(5,7,8,1) -59.9892 -DE/DX = 0.0 !
! D5 D(5,7,8,2) 60.0136 -DE/DX = 0.0 !
! D6 D(5,7,8,3) -179.9864 -DE/DX = 0.0 !
! D7 D(6,7,8,1) 60.0135 -DE/DX = 0.0 !
! D8 D(6,7,8,2) -179.9837 -DE/DX = 0.0 !
! D9 D(6,7,8,3) -59.9837 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency output information
Low frequencies --- 0.0002 0.0016 0.0018 17.2632 22.6336 39.3248 Low frequencies --- 265.9469 632.3805 639.0805
IR spectrum
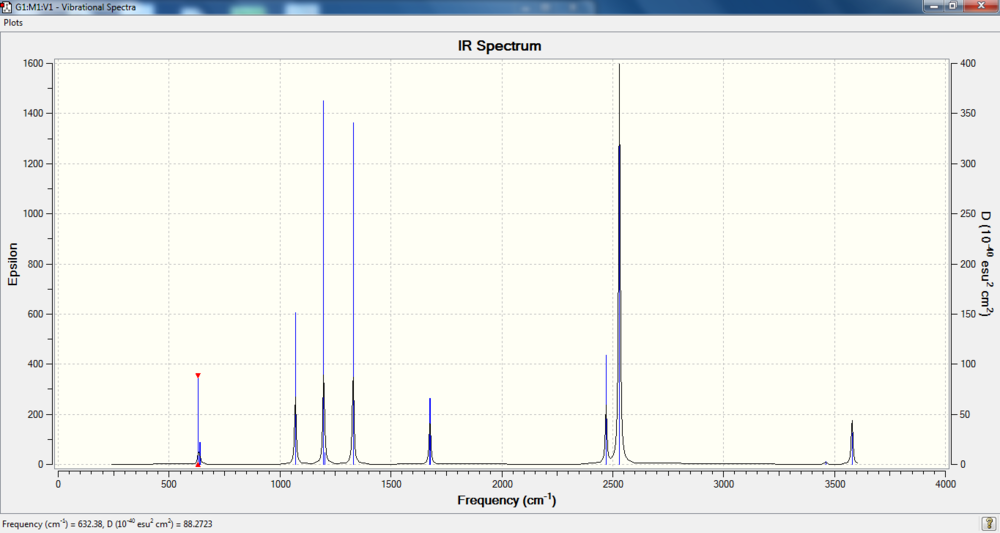
Bond information
| Bond Distance N-B | 1.66803 Å |
| Bond Distance N-H | 1.01861 Å |
| Bond Distance B-H | 1.21007 Å |
| Bond Angle H-N-B | 111.029 |
| Bond Angle H-B-N | 104.600 |
| Bond Angle H-N-H | 107.870 |
| Bond Angle H-B-H | 113.875 |
Energy calculation
E(NH3) = -56.55776856 a.u.
E(BH3) = -26.61532374 a.u.
E(NH3BH3) = -83.22468918 a.u.
ΔE = E(NH3BH3)-[E(NH3)-E(BH3)] = -83.22468918-[(-56.55776856)+(-26.46226433)] = -0.05159688 a.u. = -135.4676084 kJ/mol
File link
Optimization:File:WENMIN NH3BH3 OPT 631G DP.LOG
Frequency:File:WENMIN NH3BH3 FREQ.LOG
Week 2: Mini Project Investigating aromaticity
Benzene
The optimization, frequency and NBO calculation of benzene molecule was carried out by the HPC.
Molecule
Benzene molecule |
Summary
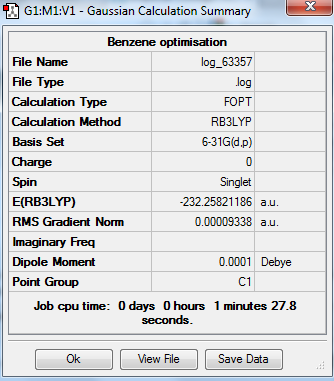
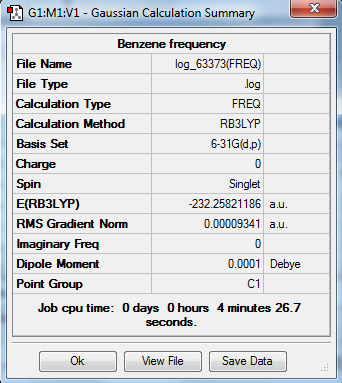
Output information
Optimization output information
Item Value Threshold Converged?
Maximum Force 0.000204 0.000450 YES
RMS Force 0.000084 0.000300 YES
Maximum Displacement 0.000870 0.001800 YES
RMS Displacement 0.000313 0.001200 YES
Predicted change in Energy=-4.983462D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0001 !
! R2 R(1,6) 1.3961 -DE/DX = 0.0002 !
! R3 R(1,7) 1.0861 -DE/DX = 0.0002 !
! R4 R(2,3) 1.3961 -DE/DX = 0.0002 !
! R5 R(2,8) 1.0861 -DE/DX = 0.0002 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0001 !
! R7 R(3,9) 1.0861 -DE/DX = 0.0002 !
! R8 R(4,5) 1.3961 -DE/DX = 0.0002 !
! R9 R(4,10) 1.0861 -DE/DX = 0.0002 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0861 -DE/DX = 0.0002 !
! R12 R(6,12) 1.0861 -DE/DX = 0.0002 !
! A1 A(2,1,6) 119.9996 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.9968 -DE/DX = 0.0 !
! A3 A(6,1,7) 120.0036 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0036 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.9916 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.0048 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.9967 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0101 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9932 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.9996 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9891 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0113 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.004 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0048 -DE/DX = 0.0 !
! A15 A(6,5,11) 119.9912 -DE/DX = 0.0 !
! A16 A(1,6,5) 119.9965 -DE/DX = 0.0 !
! A17 A(1,6,12) 120.0072 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9963 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.0059 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 180.0021 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -180.0099 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0019 -DE/DX = 0.0 !
! D5 D(2,1,6,5) -0.0055 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.9972 -DE/DX = 0.0 !
! D7 D(7,1,6,5) -180.0016 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.0068 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0119 -DE/DX = 0.0 !
! D10 D(1,2,3,9) 180.0087 -DE/DX = 0.0 !
! D11 D(8,2,3,4) 180.0039 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0007 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0064 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0058 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9968 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0026 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.005 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 180.0061 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -180.0057 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.0055 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.011 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 180.0027 -DE/DX = 0.0 !
! D23 D(11,5,6,1) 179.9999 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0085 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency output information
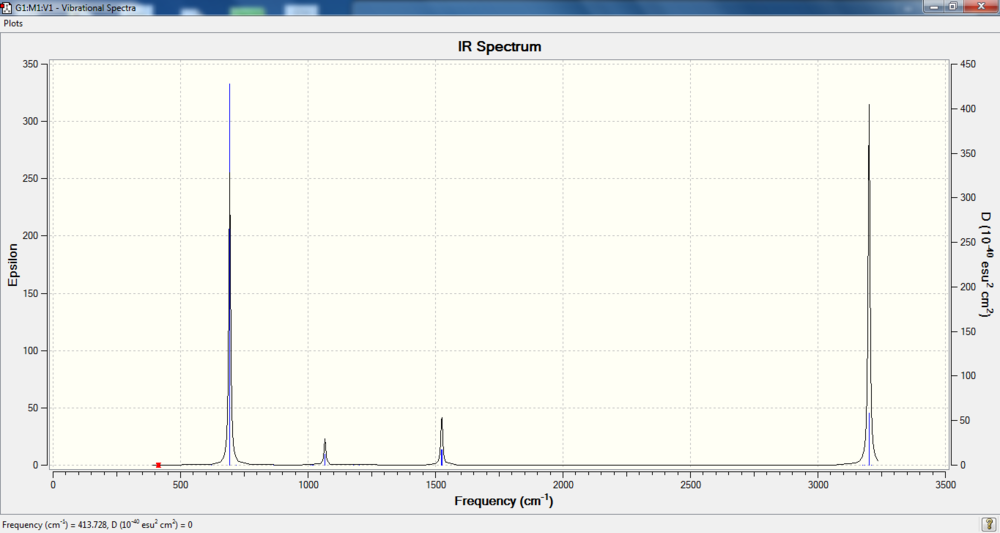
Low frequencies --- -14.2155 -2.6894 -0.0006 -0.0003 -0.0002 10.0093 Low frequencies --- 413.7279 414.5534 621.0444
NBO analysis
Natural populations
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
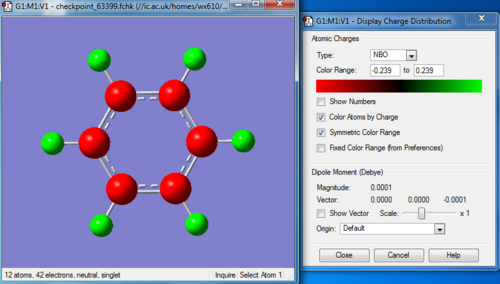
C 1 -0.23853 1.99910 4.22612 0.01331 6.23853
C 2 -0.23854 1.99910 4.22613 0.01331 6.23854
C 3 -0.23854 1.99910 4.22613 0.01331 6.23854
C 4 -0.23853 1.99910 4.22612 0.01331 6.23853
C 5 -0.23854 1.99910 4.22613 0.01331 6.23854
C 6 -0.23855 1.99910 4.22613 0.01331 6.23855
H 7 0.23854 0.00000 0.76003 0.00144 0.76146
H 8 0.23854 0.00000 0.76003 0.00144 0.76146
H 9 0.23854 0.00000 0.76002 0.00144 0.76146
H 10 0.23854 0.00000 0.76003 0.00144 0.76146
H 11 0.23854 0.00000 0.76003 0.00144 0.76146
H 12 0.23855 0.00000 0.76002 0.00144 0.76145
=======================================================================
* Total * 0.00000 11.99462 29.91691 0.08847 42.00000
Bond orbital
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98096) BD ( 1) C 1 - C 2
( 50.00%) 0.7071* C 1 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.8037
0.0163 -0.0206 -0.0317 0.0003 0.0000
0.0015 0.0000 0.0000 0.0165 -0.0109
( 50.00%) 0.7071* C 2 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.8037
-0.0163 -0.0205 -0.0317 -0.0002 0.0000
-0.0015 0.0000 0.0000 0.0165 -0.0109
2. (1.98097) BD ( 1) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.4196
-0.0357 0.6857 -0.0017 -0.0006 0.0000
-0.0135 0.0000 0.0000 -0.0096 -0.0109
( 50.00%) 0.7071* C 6 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.3840
-0.0193 -0.7063 -0.0300 0.0006 0.0000
-0.0151 0.0000 0.0000 -0.0069 -0.0109
3. (1.66532) BD ( 2) C 1 - C 6
( 50.00%) 0.7071* C 1 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 0.0006 0.0000 0.9997 -0.0133
0.0000 0.0002 0.0195 0.0000 0.0000
( 50.00%) 0.7071* C 6 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 0.0006 0.0000 0.9997 -0.0133
0.0000 0.0170 -0.0096 0.0000 0.0000
4. (1.98305) BD ( 1) C 1 - H 7
( 62.04%) 0.7876* C 1 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.4194
-0.0073 0.7265 -0.0126 -0.0003 0.0000
-0.0144 0.0000 0.0000 0.0083 0.0105
( 37.96%) 0.6161* H 7 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0114 -0.0197 0.0000
5. (1.98097) BD ( 1) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.4197
0.0357 0.6857 -0.0017 -0.0004 0.0000
0.0135 0.0000 0.0000 -0.0096 -0.0109
( 50.00%) 0.7071* C 3 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.3841
0.0193 -0.7063 -0.0300 0.0004 0.0000
0.0151 0.0000 0.0000 -0.0069 -0.0109
6. (1.66534) BD ( 2) C 2 - C 3
( 50.00%) 0.7071* C 2 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0002 0.0195 0.0000 0.0000
( 50.00%) 0.7071* C 3 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0002
0.0000 0.0007 0.0000 0.9997 -0.0133
0.0000 -0.0170 -0.0096 0.0000 0.0000
7. (1.98305) BD ( 1) C 2 - H 8
( 62.04%) 0.7876* C 2 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 -0.4193
0.0073 0.7265 -0.0126 -0.0006 0.0000
0.0144 0.0000 0.0000 0.0083 0.0105
( 37.96%) 0.6161* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 0.0114 -0.0197 0.0000
8. (1.98096) BD ( 1) C 3 - C 4
( 50.00%) 0.7071* C 3 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.3840
0.0193 0.7064 0.0300 -0.0006 0.0000
-0.0151 0.0000 0.0000 -0.0069 -0.0109
( 50.00%) 0.7071* C 4 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.4197
0.0357 -0.6858 0.0017 0.0005 0.0000
-0.0135 0.0000 0.0000 -0.0096 -0.0109
9. (1.98305) BD ( 1) C 3 - H 9
( 62.04%) 0.7876* C 3 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.8388
-0.0146 0.0000 0.0000 0.0002 0.0000
0.0000 0.0000 0.0000 0.0166 -0.0105
( 37.96%) 0.6161* H 9 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0228 0.0000 0.0000
10. (1.98097) BD ( 1) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 -0.8037
-0.0164 0.0206 0.0317 -0.0002 0.0000
0.0015 0.0000 0.0000 0.0165 -0.0109
( 50.00%) 0.7071* C 5 s( 35.20%)p 1.84( 64.76%)d 0.00( 0.04%)
-0.0001 0.5933 -0.0079 0.0006 0.8037
0.0164 0.0206 0.0317 0.0003 0.0000
-0.0015 0.0000 0.0000 0.0165 -0.0109
11. (1.66533) BD ( 2) C 4 - C 5
( 50.00%) 0.7071* C 4 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0006 0.0000 0.9997 -0.0133
0.0000 -0.0168 -0.0099 0.0000 0.0000
( 50.00%) 0.7071* C 5 s( 0.00%)p 1.00( 99.96%)d 0.00( 0.04%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 0.0006 0.0000 0.9997 -0.0133
0.0000 0.0168 -0.0099 0.0000 0.0000
12. (1.98305) BD ( 1) C 4 - H 10
( 62.04%) 0.7876* C 4 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 0.4195
-0.0073 0.7264 -0.0126 -0.0003 0.0000
0.0144 0.0000 0.0000 -0.0083 -0.0105
( 37.96%) 0.6161* H 10 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 -0.0114 -0.0197 0.0000
13. (1.98096) BD ( 1) C 5 - C 6
( 50.00%) 0.7071* C 5 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 -0.4197
-0.0357 -0.6857 0.0017 0.0003 0.0000
0.0135 0.0000 0.0000 -0.0096 -0.0109
( 50.00%) 0.7071* C 6 s( 35.19%)p 1.84( 64.77%)d 0.00( 0.04%)
-0.0001 0.5932 -0.0078 0.0006 0.3841
-0.0193 0.7063 0.0300 -0.0003 0.0000
0.0151 0.0000 0.0000 -0.0069 -0.0109
14. (1.98305) BD ( 1) C 5 - H 11
( 62.04%) 0.7876* C 5 s( 29.57%)p 2.38( 70.39%)d 0.00( 0.04%)
-0.0003 0.5437 0.0126 -0.0010 -0.4194
0.0073 0.7265 -0.0126 -0.0006 0.0000
-0.0144 0.0000 0.0000 -0.0083 -0.0105
( 37.96%) 0.6161* H 11 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0014 0.0114 -0.0197 0.0000
15. (1.98305) BD ( 1) C 6 - H 12
( 62.04%) 0.7876* C 6 s( 29.58%)p 2.38( 70.39%)d 0.00( 0.04%)
0.0003 -0.5437 -0.0126 0.0010 0.8388
-0.0146 -0.0001 0.0000 0.0003 0.0000
0.0000 0.0000 0.0000 -0.0166 0.0105
( 37.96%) 0.6161* H 12 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0014 -0.0228 0.0000 0.0000
16. (1.99911) CR ( 1) C 1 s(100.00%)p 0.00( 0.00%)
1.0000 0.0002 0.0000 0.0000 -0.0001
0.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
Second order perturbation theory analysis of Fock Matrix in NBO basis
I only selected the ones whose E2 value larger than 20 kcal/mol.
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
within unit 1 3. BD ( 2) C 1 - C 6 /111. BD*( 2) C 2 - C 3 20.42 0.28 0.068 3. BD ( 2) C 1 - C 6 /116. BD*( 2) C 4 - C 5 20.42 0.28 0.068 6. BD ( 2) C 2 - C 3 /108. BD*( 2) C 1 - C 6 20.42 0.28 0.068 6. BD ( 2) C 2 - C 3 /116. BD*( 2) C 4 - C 5 20.42 0.28 0.068 11. BD ( 2) C 4 - C 5 /108. BD*( 2) C 1 - C 6 20.42 0.28 0.068 11. BD ( 2) C 4 - C 5 /111. BD*( 2) C 2 - C 3 20.42 0.28 0.068
Natural bond orbital
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C6H6)
1. BD ( 1) C 1 - C 2 1.98096 -0.68186 110(g),107(g),114(v),120(v)
43(v),73(v),109(g),112(g)
72(v),42(v)
2. BD ( 1) C 1 - C 6 1.98097 -0.68199 106(g),118(g),119(v),112(v)
33(v),63(v),120(g),109(g)
62(v),32(v)
3. BD ( 2) C 1 - C 6 1.66532 -0.23794 116(v),111(v),65(v),35(v)
4. BD ( 1) C 1 - H 7 1.98305 -0.51235 118(v),110(v),72(v),32(v)
107(g),106(g)
5. BD ( 1) C 2 - C 3 1.98097 -0.68202 106(g),113(g),109(v),117(v)
23(v),53(v),114(g),112(g)
52(v),22(v)
6. BD ( 2) C 2 - C 3 1.66534 -0.23795 108(v),116(v),55(v),25(v)
7. BD ( 1) C 2 - H 8 1.98305 -0.51234 113(v),107(v),42(v),22(v)
110(g),106(g)
8. BD ( 1) C 3 - C 4 1.98096 -0.68183 115(g),110(g),112(v),119(v)
63(v),33(v),114(g),117(g)
32(v),62(v)
9. BD ( 1) C 3 - H 9 1.98305 -0.51236 106(v),115(v),32(v),52(v)
110(g),113(g)
10. BD ( 1) C 4 - C 5 1.98097 -0.68202 118(g),113(g),114(v),120(v)
73(v),43(v),117(g),119(g)
42(v),72(v)
11. BD ( 2) C 4 - C 5 1.66533 -0.23794 108(v),111(v),45(v),75(v)
12. BD ( 1) C 4 - H 10 1.98305 -0.51236 118(v),110(v),62(v),42(v)
115(g),113(g)
13. BD ( 1) C 5 - C 6 1.98096 -0.68186 115(g),107(g),109(v),117(v)
53(v),23(v),120(g),119(g)
22(v),52(v)
14. BD ( 1) C 5 - H 11 1.98305 -0.51234 113(v),107(v),52(v),72(v)
115(g),118(g)
15. BD ( 1) C 6 - H 12 1.98305 -0.51236 106(v),115(v),22(v),62(v)
107(g),118(g)
16. CR ( 1) C 1 1.99911 -10.04057 73(v),33(v),110(v),118(v)
120(v),112(v)
17. CR ( 1) C 2 1.99911 -10.04056 43(v),23(v),113(v),107(v)
114(v),109(v)
18. CR ( 1) C 3 1.99911 -10.04057 33(v),53(v),106(v),115(v)
112(v),117(v)
19. CR ( 1) C 4 1.99911 -10.04056 63(v),43(v),118(v),110(v)
119(v),114(v)
20. CR ( 1) C 5 1.99911 -10.04056 53(v),73(v),113(v),107(v)
117(v),120(v)
21. CR ( 1) C 6 1.99911 -10.04057 23(v),63(v),106(v),115(v)
109(v),119(v)
22. RY*( 1) C 1 0.00482 1.27866
IR spectrum
MOs of benzene
Charge distribution
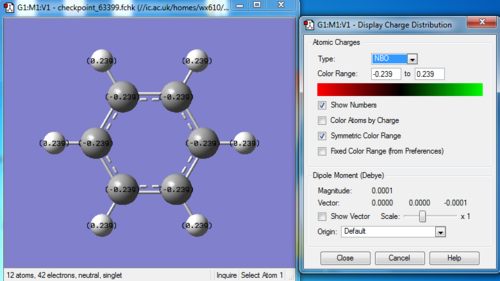
Bond information
| Bond Distance C-H | ~1.08610 Å |
| Bond Distance C1-C2 | 1.39628 Å |
| Bond Distance C2-C3 | 1.39608 Å |
| Bond Distance C3-C4 | 1.39630 Å |
| Bond Distance C4-C5 | 1.39628 Å |
| Bond Distance C5-C6 | 1.39628 Å |
| Bond Distance C6-C1 | 1.39610 Å |
| Average bond Distance C-C | ~1.39622 Å |
| Bond Angle H-C-C | ~120.000 |
| Bond Angle C-C-C | ~120.000 |
File link
Optimization:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20580
Frequency:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20581
NBO:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20581
Boratabenzene
The optimization, frequency and NBO calculation of boratabenzene molecule was carried out by the HPC.
Molecule
Boratabenzene molecule |
Summary
Output information
Optimization output information
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000068 0.000300 YES
Maximum Displacement 0.000888 0.001800 YES
RMS Displacement 0.000326 0.001200 YES
Predicted change in Energy=-6.533587D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4053 -DE/DX = -0.0001 !
! R2 R(1,5) 1.4053 -DE/DX = -0.0001 !
! R3 R(1,6) 1.0916 -DE/DX = -0.0001 !
! R4 R(2,3) 1.3989 -DE/DX = 0.0 !
! R5 R(2,7) 1.0968 -DE/DX = 0.0001 !
! R6 R(3,8) 1.097 -DE/DX = -0.0001 !
! R7 R(3,12) 1.5138 -DE/DX = 0.0001 !
! R8 R(4,5) 1.3989 -DE/DX = 0.0 !
! R9 R(4,10) 1.097 -DE/DX = -0.0001 !
! R10 R(4,12) 1.5137 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0968 -DE/DX = 0.0001 !
! R12 R(9,12) 1.2185 -DE/DX = 0.0 !
! A1 A(2,1,5) 120.4509 -DE/DX = -0.0001 !
! A2 A(2,1,6) 119.7734 -DE/DX = 0.0001 !
! A3 A(5,1,6) 119.7757 -DE/DX = 0.0001 !
! A4 A(1,2,3) 122.1382 -DE/DX = 0.0001 !
! A5 A(1,2,7) 117.4353 -DE/DX = 0.0 !
! A6 A(3,2,7) 120.4265 -DE/DX = -0.0002 !
! A7 A(2,3,8) 115.9542 -DE/DX = 0.0001 !
! A8 A(2,3,12) 120.0809 -DE/DX = -0.0001 !
! A9 A(8,3,12) 123.9649 -DE/DX = -0.0001 !
! A10 A(5,4,10) 115.9498 -DE/DX = 0.0001 !
! A11 A(5,4,12) 120.081 -DE/DX = -0.0001 !
! A12 A(10,4,12) 123.9692 -DE/DX = -0.0001 !
! A13 A(1,5,4) 122.1395 -DE/DX = 0.0001 !
! A14 A(1,5,11) 117.4369 -DE/DX = 0.0 !
! A15 A(4,5,11) 120.4236 -DE/DX = -0.0002 !
! A16 A(3,12,4) 115.1095 -DE/DX = 0.0 !
! A17 A(3,12,9) 122.4414 -DE/DX = 0.0 !
! A18 A(4,12,9) 122.4491 -DE/DX = 0.0 !
! D1 D(5,1,2,3) 0.0065 -DE/DX = 0.0 !
! D2 D(5,1,2,7) -180.0034 -DE/DX = 0.0 !
! D3 D(6,1,2,3) 180.0061 -DE/DX = 0.0 !
! D4 D(6,1,2,7) -0.0039 -DE/DX = 0.0 !
! D5 D(2,1,5,4) 0.0007 -DE/DX = 0.0 !
! D6 D(2,1,5,11) -180.0039 -DE/DX = 0.0 !
! D7 D(6,1,5,4) -179.9988 -DE/DX = 0.0 !
! D8 D(6,1,5,11) -0.0035 -DE/DX = 0.0 !
! D9 D(1,2,3,8) -180.0066 -DE/DX = 0.0 !
! D10 D(1,2,3,12) -0.0085 -DE/DX = 0.0 !
! D11 D(7,2,3,8) 0.0036 -DE/DX = 0.0 !
! D12 D(7,2,3,12) 180.0018 -DE/DX = 0.0 !
! D13 D(2,3,12,4) 0.0035 -DE/DX = 0.0 !
! D14 D(2,3,12,9) 180.0026 -DE/DX = 0.0 !
! D15 D(8,3,12,4) 180.0015 -DE/DX = 0.0 !
! D16 D(8,3,12,9) 0.0006 -DE/DX = 0.0 !
! D17 D(10,4,5,1) 179.9969 -DE/DX = 0.0 !
! D18 D(10,4,5,11) 0.0017 -DE/DX = 0.0 !
! D19 D(12,4,5,1) -0.0055 -DE/DX = 0.0 !
! D20 D(12,4,5,11) -180.0007 -DE/DX = 0.0 !
! D21 D(5,4,12,3) 0.0033 -DE/DX = 0.0 !
! D22 D(5,4,12,9) 180.0042 -DE/DX = 0.0 !
! D23 D(10,4,12,3) -179.9993 -DE/DX = 0.0 !
! D24 D(10,4,12,9) 0.0016 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency output information
Low frequencies --- -13.1286 0.0003 0.0005 0.0006 15.0635 18.1734 Low frequencies --- 371.3453 404.2332 565.2514
NBO analysis
Natural populations
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.33984 1.99907 4.32693 0.01384 6.33984
C 2 -0.25043 1.99910 4.23720 0.01412 6.25043
C 3 -0.58789 1.99901 4.57710 0.01178 6.58789
C 4 -0.58795 1.99901 4.57716 0.01178 6.58795
C 5 -0.25041 1.99910 4.23719 0.01412 6.25041
H 6 0.18564 0.00000 0.81237 0.00200 0.81436
H 7 0.17906 0.00000 0.81832 0.00262 0.82094
H 8 0.18380 0.00000 0.81402 0.00218 0.81620
H 9 -0.09642 0.00000 1.09588 0.00054 1.09642
H 10 0.18380 0.00000 0.81402 0.00218 0.81620
H 11 0.17906 0.00000 0.81832 0.00262 0.82094
B 12 0.20160 1.99906 2.78774 0.01160 4.79840
=======================================================================
* Total * -1.00000 11.99436 29.91625 0.08939 42.00000
Bond orbital
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.97970) BD ( 1) C 1 - C 2
( 50.04%) 0.7074* C 1 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 0.7062
0.0327 0.3754 -0.0141 0.0000 0.0000
0.0137 0.0000 0.0000 0.0078 -0.0107
( 49.96%) 0.7068* C 2 s( 35.50%)p 1.82( 64.46%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 -0.6874
-0.0034 -0.4135 -0.0325 0.0000 0.0000
0.0146 0.0000 0.0000 0.0081 -0.0107
2. (1.97971) BD ( 1) C 1 - C 5
( 50.04%) 0.7074* C 1 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 -0.7063
-0.0327 0.3752 -0.0141 0.0000 0.0000
-0.0137 0.0000 0.0000 0.0078 -0.0107
( 49.96%) 0.7068* C 5 s( 35.51%)p 1.82( 64.45%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 0.6875
0.0034 -0.4133 -0.0325 0.0000 0.0000
-0.0146 0.0000 0.0000 0.0081 -0.0107
3. (1.98507) BD ( 1) C 1 - H 6
( 59.44%) 0.7710* C 1 s( 28.22%)p 2.54( 71.74%)d 0.00( 0.04%)
0.0004 -0.5311 -0.0116 0.0020 -0.0001
0.0000 0.8469 -0.0076 0.0000 0.0000
0.0000 0.0000 0.0000 0.0178 0.0110
( 40.56%) 0.6369* H 6 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0011 0.0000 -0.0217 0.0000
4. (1.98270) BD ( 1) C 2 - C 3
( 50.77%) 0.7125* C 2 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 0.0573
0.0311 0.7869 0.0164 0.0000 0.0000
0.0020 0.0000 0.0000 -0.0150 -0.0098
( 49.23%) 0.7017* C 3 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 -0.0025
0.0269 -0.8201 -0.0353 -0.0001 0.0000
-0.0006 0.0000 0.0000 -0.0173 -0.0123
5. (1.76868) BD ( 2) C 2 - C 3
( 48.13%) 0.6938* C 2 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9996 -0.0213
0.0000 -0.0031 0.0171 0.0000 0.0000
( 51.87%) 0.7202* C 3 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 -0.0001 0.0000 0.9998 -0.0054
0.0000 -0.0016 -0.0185 0.0000 0.0000
6. (1.98570) BD ( 1) C 2 - H 7
( 59.32%) 0.7702* C 2 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
-0.0003 0.5182 0.0133 -0.0012 0.7228
-0.0089 -0.4562 0.0100 0.0000 0.0000
-0.0177 0.0000 0.0000 0.0069 -0.0111
( 40.68%) 0.6378* H 7 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0026 -0.0187 0.0116 0.0000
7. (1.98420) BD ( 1) C 3 - H 8
( 59.41%) 0.7708* C 3 s( 25.39%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 0.7906
-0.0003 0.3472 0.0088 0.0000 0.0000
0.0111 0.0000 0.0000 0.0149 -0.0119
( 40.59%) 0.6371* H 8 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 -0.0192 -0.0100 0.0000
8. (1.96996) BD ( 1) C 3 - B 12
( 66.70%) 0.8167* C 3 s( 42.03%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 -0.6481 -0.0158 -0.0012 0.6117
-0.0293 -0.4522 -0.0090 0.0000 0.0000
0.0059 0.0000 0.0000 -0.0041 0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.53%)d 0.00( 0.08%)
0.0000 -0.5779 0.0059 -0.0048 -0.7057
-0.0393 0.4070 -0.0096 0.0000 0.0000
0.0230 0.0000 0.0000 -0.0082 0.0133
9. (1.98270) BD ( 1) C 4 - C 5
( 49.23%) 0.7017* C 4 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 0.0027
-0.0269 -0.8202 -0.0353 0.0000 0.0000
0.0006 0.0000 0.0000 -0.0173 -0.0123
( 50.77%) 0.7125* C 5 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 -0.0575
-0.0311 0.7869 0.0164 0.0000 0.0000
-0.0020 0.0000 0.0000 -0.0150 -0.0098
10. (1.76862) BD ( 2) C 4 - C 5
( 51.87%) 0.7202* C 4 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9998 -0.0054
0.0000 0.0016 -0.0185 0.0000 0.0000
( 48.13%) 0.6937* C 5 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9996 -0.0213
0.0000 0.0031 0.0171 0.0000 0.0000
11. (1.98420) BD ( 1) C 4 - H 10
( 59.41%) 0.7708* C 4 s( 25.38%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 -0.7907
0.0003 0.3469 0.0088 0.0000 0.0000
-0.0111 0.0000 0.0000 0.0150 -0.0119
( 40.59%) 0.6371* H 10 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 0.0192 -0.0100 0.0000
12. (1.96997) BD ( 1) C 4 - B 12
( 66.70%) 0.8167* C 4 s( 42.04%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 0.6482 0.0158 0.0012 0.6115
-0.0293 0.4524 0.0090 0.0000 0.0000
0.0059 0.0000 0.0000 0.0041 -0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 0.5779 -0.0059 0.0048 -0.7056
-0.0393 -0.4071 0.0096 0.0000 0.0000
0.0230 0.0000 0.0000 0.0082 -0.0133
13. (1.98570) BD ( 1) C 5 - H 11
( 59.32%) 0.7702* C 5 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
0.0003 -0.5182 -0.0133 0.0012 0.7227
-0.0089 0.4563 -0.0100 0.0000 0.0000
-0.0177 0.0000 0.0000 -0.0069 0.0111
( 40.68%) 0.6378* H 11 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0026 -0.0187 -0.0116 0.0000
14. (1.98604) BD ( 1) H 9 - B 12
( 55.09%) 0.7422* H 9 s( 99.97%)p 0.00( 0.03%)
0.9998 0.0001 0.0000 -0.0180 0.0000
( 44.91%) 0.6702* B 12 s( 33.16%)p 2.01( 66.78%)d 0.00( 0.06%)
-0.0005 0.5758 0.0069 -0.0060 -0.0001
0.0000 0.8172 -0.0016 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0213 -0.0105
15. (1.99907) CR ( 1) C 1 s(100.00%)p 0.00( 0.00%)
1.0000 0.0003 0.0000 0.0000 0.0000
0.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
Second order perturbation theory analysis of Fock Matrix in NBO basis
I only selected the ones whose E2 value is larger than 20kcal/mol.
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
within unit 1 5. BD ( 2) C 2 - C 3 / 21. LP ( 1) C 1 37.20 0.13 0.083 5. BD ( 2) C 2 - C 3 / 22. LP*( 1) B 12 24.27 0.25 0.077 10. BD ( 2) C 4 - C 5 / 21. LP ( 1) C 1 37.21 0.13 0.083 10. BD ( 2) C 4 - C 5 / 22. LP*( 1) B 12 24.28 0.25 0.077 21. LP ( 1) C 1 /111. BD*( 2) C 2 - C 3 71.49 0.15 0.106 21. LP ( 1) C 1 /116. BD*( 2) C 4 - C 5 71.51 0.15 0.106 22. LP*( 1) B 12 /111. BD*( 2) C 2 - C 3 174.83 0.02 0.086 22. LP*( 1) B 12 /116. BD*( 2) C 4 - C 5 174.95 0.02 0.086
Natural bond orbital
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6B)
1. BD ( 1) C 1 - C 2 1.97970 -0.46973 110(g),108(g),119(v),113(v)
64(v),43(v),109(g),112(g)
2. BD ( 1) C 1 - C 5 1.97971 -0.46976 115(g),107(g),112(v),117(v)
34(v),53(v),109(g),119(g)
3. BD ( 1) C 1 - H 6 1.98507 -0.31744 115(v),110(v),63(v),33(v)
108(g),107(g)
4. BD ( 1) C 2 - C 3 1.98270 -0.46498 114(g),107(g),109(v),112(g)
24(v),120(v),113(g),98(v)
23(v)
5. BD ( 2) C 2 - C 3 1.76868 -0.02908 21(v),22(v),25(v),100(v)
111(g)
6. BD ( 1) C 2 - H 7 1.98570 -0.31412 108(v),114(v),23(v),43(v)
110(g)
7. BD ( 1) C 3 - H 8 1.98420 -0.28849 107(v),114(g),33(v),118(v)
97(v),110(g)
8. BD ( 1) C 3 - B 12 1.96996 -0.31775 110(g),112(v),117(v),113(g)
34(v),33(v),81(v),54(v)
118(g)
9. BD ( 1) C 4 - C 5 1.98270 -0.46495 118(g),108(g),109(v),119(g)
24(v),120(v),117(g),98(v)
23(v)
10. BD ( 2) C 4 - C 5 1.76862 -0.02907 21(v),22(v),25(v),100(v)
116(g)
11. BD ( 1) C 4 - H 10 1.98420 -0.28848 108(v),118(g),63(v),114(v)
97(v),115(g)
12. BD ( 1) C 4 - B 12 1.96997 -0.31779 115(g),119(v),113(v),117(g)
64(v),63(v),89(v),44(v)
114(g)
13. BD ( 1) C 5 - H 11 1.98570 -0.31412 107(v),118(v),23(v),53(v)
115(g)
14. BD ( 1) H 9 - B 12 1.98604 -0.17253 115(v),110(v),53(v),43(v)
15. CR ( 1) C 1 1.99907 -9.82827 64(v),34(v),115(v),110(v)
119(v),112(v),37(v),67(v)
63(v),33(v)
16. CR ( 1) C 2 1.99910 -9.83478 44(v),24(v),114(v),109(v)
108(v),113(v)
17. CR ( 1) C 3 1.99902 -9.79408 34(v),98(v),114(g),107(v)
112(v),97(v)
18. CR ( 1) C 4 1.99902 -9.79407 64(v),98(v),118(g),108(v)
119(v),97(v)
19. CR ( 1) C 5 1.99910 -9.83478 54(v),24(v),118(v),109(v)
107(v),117(v)
20. CR ( 1) B 12 1.99907 -6.36942 117(v),113(v),115(v),110(v)
53(v),43(v)
21. LP ( 1) C 1 1.14688 0.09689 116(v),111(v),25(g),66(v)
36(v),35(v),65(v)
22. LP*( 1) B 12 0.57262 0.22267 116(v),111(v),100(g),57(v)
47(v)
23. RY*( 1) C 1 0.00435 1.45200
IR spectrum
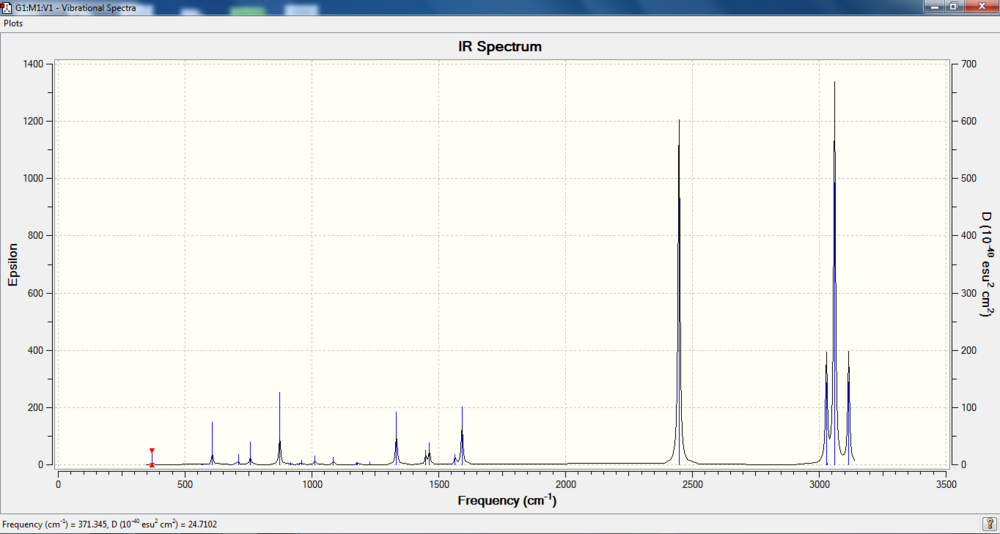
Charge distribution
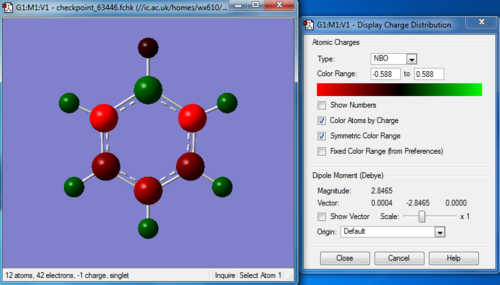
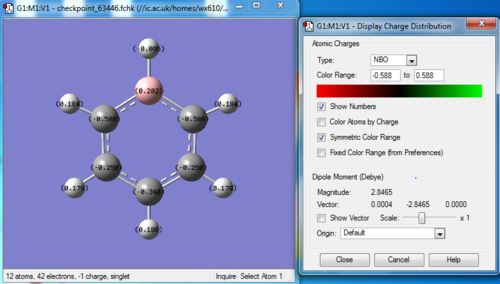
Bond information
| Bond Distance B-H | 1.21848 Å |
| Bond Distance C1(4)-H | 1.09698 Å |
| Bond Distance C2(5)-H | 1.09677 Å |
| Bond Distance C3-H | 1.09165 Å |
| Bond Distance B-C3 | 1.51383 Å |
| Bond Distance B-C4 | 1.51374 Å |
| Bond Distance C3-C2 | 1.39887 Å |
| Bond Distance C2-C1 | 1.40532 Å |
| Bond Distance C1-C5 | 1.40528 Å |
| Bond Distance C5-C4 | 1.39890 Å |
| Bond Angle H-B-C | ~122.445 |
| Bond Angle H-C-B | ~123.967 |
| Bond Angle B-C-C | 120.081 |
| Bond Angle C3(4)-C2(5)-C1 | 122.138 |
| Bond Angle C2-C1-C5 | 120.451 |
File link
Optimization:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20619
Frequency:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20622
NBO:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20624
Pyridinium
The optimization, frequency and NBO calculation of pyridinium molecule was carried out by the HPC.
Molecule
Pyridinium molecule |
Summary

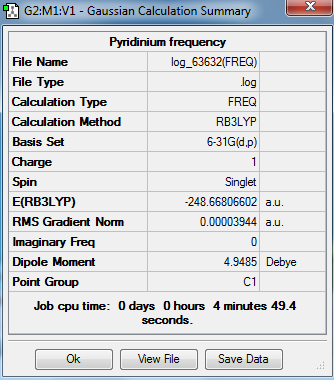
Output information
Optimization output information
Item Value Threshold Converged?
Maximum Force 0.000065 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000830 0.001800 YES
RMS Displacement 0.000176 0.001200 YES
Predicted change in Energy=-7.038332D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3988 -DE/DX = 0.0 !
! R2 R(1,5) 1.3988 -DE/DX = 0.0 !
! R3 R(1,6) 1.0852 -DE/DX = 0.0 !
! R4 R(2,3) 1.3838 -DE/DX = 0.0 !
! R5 R(2,7) 1.0835 -DE/DX = 0.0 !
! R6 R(3,8) 1.0832 -DE/DX = 0.0 !
! R7 R(3,12) 1.3524 -DE/DX = 0.0 !
! R8 R(4,5) 1.3839 -DE/DX = 0.0 !
! R9 R(4,10) 1.0832 -DE/DX = 0.0 !
! R10 R(4,12) 1.3523 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0835 -DE/DX = 0.0 !
! R12 R(9,12) 1.0169 -DE/DX = 0.0 !
! A1 A(2,1,5) 120.0614 -DE/DX = 0.0 !
! A2 A(2,1,6) 119.9688 -DE/DX = 0.0 !
! A3 A(5,1,6) 119.9698 -DE/DX = 0.0 !
! A4 A(1,2,3) 119.0799 -DE/DX = 0.0 !
! A5 A(1,2,7) 121.4985 -DE/DX = -0.0001 !
! A6 A(3,2,7) 119.4216 -DE/DX = 0.0 !
! A7 A(2,3,8) 123.9337 -DE/DX = 0.0 !
! A8 A(2,3,12) 119.2332 -DE/DX = 0.0 !
! A9 A(8,3,12) 116.8331 -DE/DX = 0.0 !
! A10 A(5,4,10) 123.9279 -DE/DX = 0.0 !
! A11 A(5,4,12) 119.2355 -DE/DX = 0.0 !
! A12 A(10,4,12) 116.8366 -DE/DX = 0.0 !
! A13 A(1,5,4) 119.0771 -DE/DX = 0.0 !
! A14 A(1,5,11) 121.503 -DE/DX = -0.0001 !
! A15 A(4,5,11) 119.42 -DE/DX = 0.0001 !
! A16 A(3,12,4) 123.3129 -DE/DX = 0.0 !
! A17 A(3,12,9) 118.343 -DE/DX = 0.0 !
! A18 A(4,12,9) 118.344 -DE/DX = 0.0 !
! D1 D(5,1,2,3) 0.0007 -DE/DX = 0.0 !
! D2 D(5,1,2,7) -180.0002 -DE/DX = 0.0 !
! D3 D(6,1,2,3) 180.0013 -DE/DX = 0.0 !
! D4 D(6,1,2,7) 0.0004 -DE/DX = 0.0 !
! D5 D(2,1,5,4) 0.0012 -DE/DX = 0.0 !
! D6 D(2,1,5,11) 180.0005 -DE/DX = 0.0 !
! D7 D(6,1,5,4) 180.0005 -DE/DX = 0.0 !
! D8 D(6,1,5,11) -0.0002 -DE/DX = 0.0 !
! D9 D(1,2,3,8) -180.0002 -DE/DX = 0.0 !
! D10 D(1,2,3,12) -0.0018 -DE/DX = 0.0 !
! D11 D(7,2,3,8) 0.0007 -DE/DX = 0.0 !
! D12 D(7,2,3,12) -180.001 -DE/DX = 0.0 !
! D13 D(2,3,12,4) 0.0012 -DE/DX = 0.0 !
! D14 D(2,3,12,9) 180.0012 -DE/DX = 0.0 !
! D15 D(8,3,12,4) -180.0003 -DE/DX = 0.0 !
! D16 D(8,3,12,9) -0.0003 -DE/DX = 0.0 !
! D17 D(10,4,5,1) 180.0005 -DE/DX = 0.0 !
! D18 D(10,4,5,11) 0.0012 -DE/DX = 0.0 !
! D19 D(12,4,5,1) -0.0018 -DE/DX = 0.0 !
! D20 D(12,4,5,11) -180.0011 -DE/DX = 0.0 !
! D21 D(5,4,12,3) 0.0006 -DE/DX = 0.0 !
! D22 D(5,4,12,9) 180.0006 -DE/DX = 0.0 !
! D23 D(10,4,12,3) -180.0015 -DE/DX = 0.0 !
! D24 D(10,4,12,9) -0.0015 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency output information
Low frequencies --- -5.3818 -0.0008 -0.0006 0.0008 11.0036 14.1046 Low frequencies --- 391.5100 404.4645 620.4186
NBO analysis
Natural populations
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.12243 1.99913 4.10943 0.01386 6.12243
C 2 -0.24103 1.99912 4.22859 0.01331 6.24103
C 3 0.07099 1.99918 3.91068 0.01916 5.92901
C 4 0.07101 1.99918 3.91066 0.01916 5.92899
C 5 -0.24103 1.99912 4.22859 0.01331 6.24103
H 6 0.29169 0.00000 0.70718 0.00113 0.70831
H 7 0.29719 0.00000 0.70178 0.00103 0.70281
H 8 0.28493 0.00000 0.71397 0.00110 0.71507
H 9 0.48278 0.00000 0.51476 0.00246 0.51722
H 10 0.28493 0.00000 0.71397 0.00110 0.71507
H 11 0.29719 0.00000 0.70178 0.00103 0.70281
N 12 -0.47622 1.99937 5.46756 0.00929 7.47622
=======================================================================
* Total * 1.00000 11.99510 29.90895 0.09595 42.00000
Bond orbital
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98249) BD ( 1) C 1 - C 2
( 49.74%) 0.7053* C 1 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 -0.3938
0.0234 0.7061 0.0290 0.0003 0.0000
-0.0169 0.0000 0.0000 -0.0060 -0.0113
( 50.26%) 0.7089* C 2 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 0.4188
0.0371 -0.6896 0.0068 -0.0002 0.0000
-0.0122 0.0000 0.0000 -0.0118 -0.0115
2. (1.98249) BD ( 1) C 1 - C 5
( 49.74%) 0.7053* C 1 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 -0.3931
0.0235 -0.7065 -0.0289 -0.0006 0.0000
0.0169 0.0000 0.0000 -0.0060 -0.0113
( 50.26%) 0.7089* C 5 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 0.4181
0.0371 0.6900 -0.0068 0.0006 0.0000
0.0122 0.0000 0.0000 -0.0119 -0.0115
3. (1.54880) BD ( 2) C 1 - C 5
( 45.73%) 0.6762* C 1 s( 0.00%)p 1.00( 99.93%)d 0.00( 0.07%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 -0.0006 0.0000 0.9997 -0.0036
0.0000 -0.0241 -0.0101 0.0000 0.0000
( 54.27%) 0.7367* C 5 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 -0.0006 0.0000 0.9997 -0.0080
0.0000 -0.0086 0.0228 0.0000 0.0000
4. (1.98141) BD ( 1) C 1 - H 6
( 64.64%) 0.8040* C 1 s( 31.07%)p 2.22( 68.90%)d 0.00( 0.03%)
-0.0003 0.5572 0.0131 -0.0007 0.8298
-0.0198 0.0004 0.0000 0.0003 0.0000
0.0000 0.0000 0.0000 0.0153 -0.0101
( 35.36%) 0.5947* H 6 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0018 -0.0242 0.0000 0.0000
5. (1.98297) BD ( 1) C 2 - C 3
( 49.58%) 0.7042* C 2 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 -0.8145
-0.0194 -0.0011 0.0320 -0.0003 0.0000
0.0047 0.0000 0.0000 0.0179 -0.0119
( 50.42%) 0.7100* C 3 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 0.7832
0.0046 0.0145 0.0331 0.0003 0.0000
-0.0053 0.0000 0.0000 0.0168 -0.0095
6. (1.61445) BD ( 2) C 2 - C 3
( 52.23%) 0.7227* C 2 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 -0.0006 0.0000 0.9997 -0.0068
0.0000 -0.0191 -0.0159 0.0000 0.0000
( 47.77%) 0.6912* C 3 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 -0.0006 0.0000 0.9995 -0.0175
0.0000 0.0196 -0.0153 0.0000 0.0000
7. (1.97822) BD ( 1) C 2 - H 7
( 64.83%) 0.8052* C 2 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
-0.0003 0.5636 0.0138 -0.0005 0.3985
-0.0072 0.7230 -0.0181 0.0006 0.0000
0.0109 0.0000 0.0000 -0.0085 -0.0099
( 35.17%) 0.5930* H 7 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0016 -0.0116 -0.0208 0.0000
8. (1.98154) BD ( 1) C 3 - H 8
( 64.26%) 0.8016* C 3 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
-0.0004 0.5780 0.0180 -0.0017 -0.4697
0.0193 0.6663 -0.0183 0.0002 0.0000
-0.0164 0.0000 0.0000 -0.0019 -0.0092
( 35.74%) 0.5978* H 8 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0018 0.0129 -0.0209 0.0000
9. (1.98861) BD ( 1) C 3 - N 12
( 36.68%) 0.6057* C 3 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5293 -0.0335 -0.0013 -0.4039
-0.0563 -0.7419 -0.0277 -0.0006 0.0000
0.0251 0.0000 0.0000 -0.0185 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6046 -0.0037 0.0006 0.3655
-0.0187 0.7071 0.0132 0.0006 0.0000
0.0107 0.0000 0.0000 -0.0059 -0.0115
10. (1.98297) BD ( 1) C 4 - C 5
( 50.42%) 0.7100* C 4 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 0.7832
0.0047 -0.0137 -0.0331 0.0003 0.0000
0.0053 0.0000 0.0000 0.0168 -0.0095
( 49.58%) 0.7042* C 5 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 -0.8145
-0.0194 0.0004 -0.0320 -0.0003 0.0000
-0.0047 0.0000 0.0000 0.0179 -0.0119
11. (1.98154) BD ( 1) C 4 - H 10
( 64.26%) 0.8016* C 4 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
0.0004 -0.5780 -0.0180 0.0017 0.4690
-0.0193 0.6667 -0.0184 0.0006 0.0000
-0.0164 0.0000 0.0000 0.0019 0.0092
( 35.74%) 0.5978* H 10 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 -0.0128 -0.0209 0.0000
12. (1.98861) BD ( 1) C 4 - N 12
( 36.68%) 0.6057* C 4 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5294 -0.0335 -0.0013 -0.4046
-0.0563 0.7415 0.0276 0.0003 0.0000
-0.0252 0.0000 0.0000 -0.0184 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6047 -0.0037 0.0006 0.3662
-0.0186 -0.7067 -0.0132 -0.0003 0.0000
-0.0107 0.0000 0.0000 -0.0058 -0.0115
13. (1.82448) BD ( 2) C 4 - N 12
( 28.55%) 0.5343* C 4 s( 0.00%)p 1.00( 99.83%)d 0.00( 0.17%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 -0.0006 0.0000 0.9991 0.0132
0.0000 -0.0102 0.0394 0.0000 0.0000
( 71.45%) 0.8453* N 12 s( 0.00%)p 1.00( 99.98%)d 0.00( 0.02%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 -0.0006 0.0000 0.9999 0.0036
0.0000 0.0128 -0.0077 0.0000 0.0000
14. (1.97822) BD ( 1) C 5 - H 11
( 64.83%) 0.8052* C 5 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
0.0003 -0.5636 -0.0138 0.0005 -0.3991
0.0072 0.7226 -0.0181 0.0003 0.0000
0.0110 0.0000 0.0000 0.0085 0.0099
( 35.17%) 0.5930* H 11 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0016 0.0116 -0.0208 0.0000
15. (1.98629) BD ( 1) H 9 - N 12
( 25.41%) 0.5041* H 9 s( 99.88%)p 0.00( 0.12%)
-0.9994 0.0064 -0.0342 0.0000 0.0000
( 74.59%) 0.8637* N 12 s( 26.81%)p 2.73( 73.16%)d 0.00( 0.02%)
0.0002 -0.5178 -0.0066 0.0013 0.8553
-0.0091 0.0004 0.0000 0.0003 0.0000
0.0000 0.0000 0.0000 -0.0115 0.0106
16. (1.99914) CR ( 1) C 1 s(100.00%)p 0.00( 0.00%)
1.0000 0.0002 0.0000 0.0000 0.0002
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
Second order perturbation theory analysis of Fock Matrix in NBO basis
I only selected the ones whose E2 value is larger than 20kcal/mol.
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
within unit 1 3. BD ( 2) C 1 - C 5 /111. BD*( 2) C 2 - C 3 16.09 0.27 0.062 3. BD ( 2) C 1 - C 5 /118. BD*( 2) C 4 - N 12 47.06 0.20 0.089 6. BD ( 2) C 2 - C 3 /108. BD*( 2) C 1 - C 5 20.71 0.30 0.073 6. BD ( 2) C 2 - C 3 /118. BD*( 2) C 4 - N 12 15.42 0.22 0.054 13. BD ( 2) C 4 - N 12 /111. BD*( 2) C 2 - C 3 20.53 0.39 0.081 118. BD*( 2) C 4 - N 12 /108. BD*( 2) C 1 - C 5 50.25 0.08 0.090 118. BD*( 2) C 4 - N 12 /111. BD*( 2) C 2 - C 3 31.24 0.07 0.068
Natural bond orbital
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6N)
1. BD ( 1) C 1 - C 2 1.98249 -0.90378 113(v),119(v),110(g),43(v)
107(g),112(g),63(v),62(v)
109(g)
2. BD ( 1) C 1 - C 5 1.98249 -0.90379 116(v),112(v),115(g),54(v)
106(g),119(g),33(v),32(v)
109(g)
3. BD ( 2) C 1 - C 5 1.54880 -0.44892 118(v),111(v),56(v),36(v)
4. BD ( 1) C 1 - H 6 1.98141 -0.71804 110(v),115(v),32(v),62(v)
107(g),106(g)
5. BD ( 1) C 2 - C 3 1.98297 -0.92650 120(v),109(v),106(g),23(v)
113(g),97(v),112(g),22(v)
114(g),96(v)
6. BD ( 2) C 2 - C 3 1.61445 -0.46666 108(v),118(v),99(v),111(g)
27(v)
7. BD ( 1) C 2 - H 7 1.97822 -0.71855 114(v),107(v),22(v),42(v)
106(g),110(g)
8. BD ( 1) C 3 - H 8 1.98154 -0.75116 117(v),106(v),32(v),110(g)
96(v)
9. BD ( 1) C 3 - N 12 1.98861 -1.06560 117(g),52(v),112(v),33(v)
116(v),54(v),110(g),120(g)
10. BD ( 1) C 4 - C 5 1.98297 -0.92647 120(v),109(v),107(g),23(v)
116(g),97(v),119(g),22(v)
117(g),96(v)
11. BD ( 1) C 4 - H 10 1.98154 -0.75116 114(v),107(v),62(v),115(g)
96(v)
12. BD ( 1) C 4 - N 12 1.98861 -1.06564 114(g),42(v),119(v),63(v)
113(v),43(v),115(g),120(g)
13. BD ( 2) C 4 - N 12 1.82448 -0.56812 111(v),108(v),44(v),66(v)
14. BD ( 1) C 5 - H 11 1.97822 -0.71856 117(v),106(v),22(v),52(v)
107(g),115(g)
15. BD ( 1) H 9 - N 12 1.98629 -0.89228 115(v),110(v),52(v),42(v)
16. CR ( 1) C 1 1.99914 -10.27397 33(v),63(v),110(v),115(v)
119(v),112(v),72(v)
17. CR ( 1) C 2 1.99913 -10.26482 23(v),43(v),42(v),113(v)
107(v),76(v),109(v),114(v)
18. CR ( 1) C 3 1.99918 -10.32332 33(v),117(v),120(v),106(v)
110(g),97(v),80(v),112(v)
19. CR ( 1) C 4 1.99918 -10.32333 63(v),114(v),120(v),107(v)
115(g),97(v),88(v),119(v)
20. CR ( 1) C 5 1.99913 -10.26483 23(v),54(v),52(v),116(v)
106(v),92(v),109(v),117(v)
21. CR ( 1) N 12 1.99937 -14.46219 54(v),43(v)
22. RY*( 1) C 1 0.00524 1.08103
IR spectrum
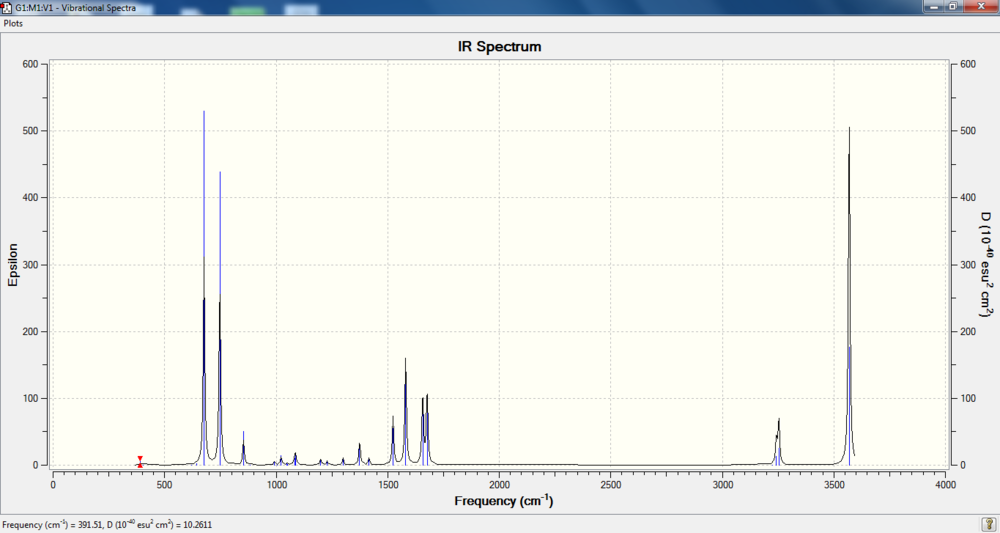
Charge distribution
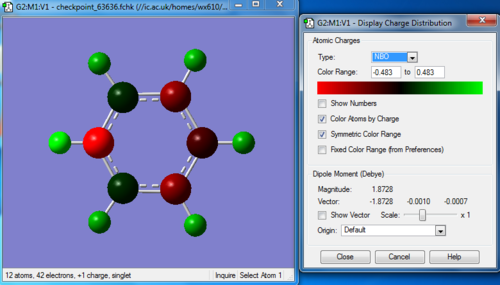
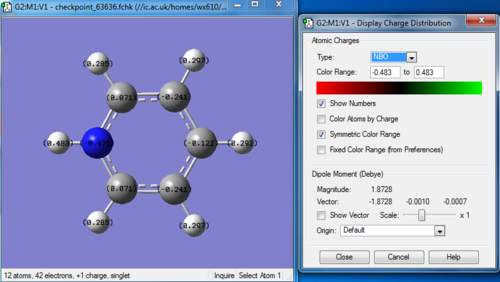
Bond information
| Bond Distance N-H | 1.01692 Å |
| Bond Distance C1(4)-H | 1.08521 Å |
| Bond Distance C2(5)-H | 1.08352 Å |
| Bond Distance C3-H | 1.08324 Å |
| Bond Distance N-C3 | 1.35237 Å |
| Bond Distance N-C4 | 1.35233 Å |
| Bond Distance C3-C2 | 1.38384 Å |
| Bond Distance C2-C1 | 1.39877 Å |
| Bond Distance C1-C5 | 1.39876 Å |
| Bond Distance C5-C4 | 1.38388 Å |
| Bond Angle H-N-C | ~118.344 |
| Bond Angle H-C-N | ~116.835 |
| Bond Angle N-C-C | ~119.235 |
| Bond Angle C3(4)-C2(5)-C1 | ~119.078 |
| Bond Angle C2-C1-C5 | 120.061 |
File link
Optimization:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20699
Frequency:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20702
NBO:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20704
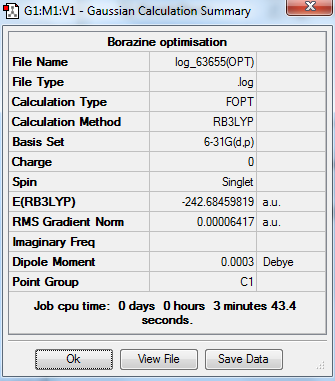
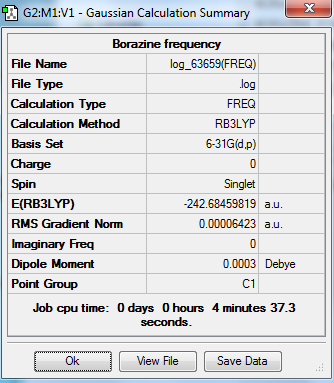
Borazine
The optimization, frequency and NBO calculation of pyridinium molecule was carried out by the HPC.
Molecule
Borazine molecule |
Summary
Output information
Optimization output information
Item Value Threshold Converged?
Maximum Force 0.000086 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.000252 0.001800 YES
RMS Displacement 0.000077 0.001200 YES
Predicted change in Energy=-9.332344D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,11) 1.1949 -DE/DX = 0.0001 !
! R2 R(2,9) 1.0097 -DE/DX = 0.0 !
! R3 R(3,12) 1.1949 -DE/DX = 0.0001 !
! R4 R(4,7) 1.0097 -DE/DX = 0.0 !
! R5 R(5,10) 1.1949 -DE/DX = 0.0001 !
! R6 R(6,8) 1.0097 -DE/DX = 0.0 !
! R7 R(7,10) 1.4307 -DE/DX = -0.0001 !
! R8 R(7,12) 1.4307 -DE/DX = -0.0001 !
! R9 R(8,10) 1.4307 -DE/DX = -0.0001 !
! R10 R(8,11) 1.4306 -DE/DX = -0.0001 !
! R11 R(9,11) 1.4307 -DE/DX = -0.0001 !
! R12 R(9,12) 1.4307 -DE/DX = -0.0001 !
! A1 A(4,7,10) 118.5641 -DE/DX = 0.0 !
! A2 A(4,7,12) 118.5579 -DE/DX = 0.0 !
! A3 A(10,7,12) 122.878 -DE/DX = 0.0 !
! A4 A(6,8,10) 118.5602 -DE/DX = 0.0 !
! A5 A(6,8,11) 118.5628 -DE/DX = 0.0 !
! A6 A(10,8,11) 122.877 -DE/DX = 0.0 !
! A7 A(2,9,11) 118.5609 -DE/DX = 0.0 !
! A8 A(2,9,12) 118.561 -DE/DX = 0.0 !
! A9 A(11,9,12) 122.8781 -DE/DX = 0.0 !
! A10 A(5,10,7) 121.4399 -DE/DX = 0.0 !
! A11 A(5,10,8) 121.437 -DE/DX = 0.0 !
! A12 A(7,10,8) 117.123 -DE/DX = 0.0 !
! A13 A(1,11,8) 121.4407 -DE/DX = 0.0 !
! A14 A(1,11,9) 121.4364 -DE/DX = 0.0 !
! A15 A(8,11,9) 117.1229 -DE/DX = 0.0 !
! A16 A(3,12,7) 121.4374 -DE/DX = 0.0 !
! A17 A(3,12,9) 121.4415 -DE/DX = 0.0 !
! A18 A(7,12,9) 117.1211 -DE/DX = 0.0 !
! D1 D(4,7,10,5) 0.0036 -DE/DX = 0.0 !
! D2 D(4,7,10,8) -179.9962 -DE/DX = 0.0 !
! D3 D(12,7,10,5) -179.9997 -DE/DX = 0.0 !
! D4 D(12,7,10,8) 0.0005 -DE/DX = 0.0 !
! D5 D(4,7,12,3) 0.0007 -DE/DX = 0.0 !
! D6 D(4,7,12,9) -179.9989 -DE/DX = 0.0 !
! D7 D(10,7,12,3) -179.996 -DE/DX = 0.0 !
! D8 D(10,7,12,9) 0.0044 -DE/DX = 0.0 !
! D9 D(6,8,10,5) 0.0003 -DE/DX = 0.0 !
! D10 D(6,8,10,7) -179.9999 -DE/DX = 0.0 !
! D11 D(11,8,10,5) 179.997 -DE/DX = 0.0 !
! D12 D(11,8,10,7) -0.0032 -DE/DX = 0.0 !
! D13 D(6,8,11,1) -0.0025 -DE/DX = 0.0 !
! D14 D(6,8,11,9) 179.9975 -DE/DX = 0.0 !
! D15 D(10,8,11,1) -179.9992 -DE/DX = 0.0 !
! D16 D(10,8,11,9) 0.0008 -DE/DX = 0.0 !
! D17 D(2,9,11,1) -0.0057 -DE/DX = 0.0 !
! D18 D(2,9,11,8) 179.9944 -DE/DX = 0.0 !
! D19 D(12,9,11,1) -179.9955 -DE/DX = 0.0 !
! D20 D(12,9,11,8) 0.0045 -DE/DX = 0.0 !
! D21 D(2,9,12,3) 0.0035 -DE/DX = 0.0 !
! D22 D(2,9,12,7) -179.9968 -DE/DX = 0.0 !
! D23 D(11,9,12,3) 179.9933 -DE/DX = 0.0 !
! D24 D(11,9,12,7) -0.007 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency output information
Low frequencies --- -7.0969 -0.0009 0.0006 0.0007 7.3516 13.0343 Low frequencies --- 288.5920 290.5614 404.2308
NBO analysis
Natural populations
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
H 1 -0.07654 0.00000 1.07585 0.00069 1.07654
H 2 0.43198 0.00000 0.56573 0.00228 0.56802
H 3 -0.07654 0.00000 1.07584 0.00069 1.07654
H 4 0.43199 0.00000 0.56573 0.00228 0.56801
H 5 -0.07654 0.00000 1.07585 0.00069 1.07654
H 6 0.43199 0.00000 0.56573 0.00228 0.56801
N 7 -1.10242 1.99943 6.09821 0.00478 8.10242
N 8 -1.10241 1.99943 6.09820 0.00478 8.10241
N 9 -1.10241 1.99943 6.09820 0.00478 8.10241
B 10 0.74699 1.99917 2.23864 0.01520 4.25301
B 11 0.74698 1.99917 2.23864 0.01520 4.25302
B 12 0.74694 1.99917 2.23868 0.01520 4.25306
=======================================================================
* Total * 0.00000 11.99579 29.93532 0.06889 42.00000
Bond orbital
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98670) BD ( 1) H 1 - B 11
( 54.03%) 0.7351* H 1 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0097 0.0165 0.0000
( 45.97%) 0.6780* B 11 s( 37.47%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.4010
0.0137 -0.6805 0.0232 0.0003 0.0000
0.0207 0.0000 0.0000 -0.0114 -0.0098
2. (1.98495) BD ( 1) H 2 - N 9
( 28.08%) 0.5299* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 0.0145 -0.0257 0.0000
( 71.92%) 0.8481* N 9 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 -0.4323
-0.0064 0.7645 0.0114 -0.0007 0.0000
0.0104 0.0000 0.0000 0.0063 0.0119
3. (1.98670) BD ( 1) H 3 - B 12
( 54.03%) 0.7351* H 3 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 -0.0192 0.0002 0.0000
( 45.97%) 0.6780* B 12 s( 37.48%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 0.7898
-0.0269 -0.0070 0.0002 0.0003 0.0000
-0.0004 0.0000 0.0000 0.0236 -0.0098
4. (1.98495) BD ( 1) H 4 - N 7
( 28.08%) 0.5299* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 -0.0010 -0.0150 -0.0254 0.0000
( 71.92%) 0.8481* N 7 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
-0.0002 0.4776 -0.0114 0.0006 0.4459
0.0066 0.7566 0.0112 -0.0003 0.0000
0.0107 0.0000 0.0000 -0.0059 -0.0119
5. (1.98670) BD ( 1) H 5 - B 10
( 54.03%) 0.7351* H 5 s( 99.96%)p 0.00( 0.04%)
0.9998 0.0002 0.0094 -0.0167 0.0000
( 45.97%) 0.6780* B 10 s( 37.47%)p 1.67( 62.46%)d 0.00( 0.07%)
-0.0006 0.6120 0.0129 -0.0016 -0.3888
0.0132 0.6875 -0.0234 -0.0006 0.0000
-0.0202 0.0000 0.0000 -0.0122 -0.0098
6. (1.98495) BD ( 1) H 6 - N 8
( 28.08%) 0.5299* H 6 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0010 -0.0295 0.0003 0.0000
( 71.92%) 0.8481* N 8 s( 22.82%)p 3.38( 77.15%)d 0.00( 0.03%)
0.0002 -0.4776 0.0114 -0.0006 0.8782
0.0131 -0.0078 -0.0001 0.0003 0.0000
0.0002 0.0000 0.0000 -0.0122 0.0119
7. (1.98438) BD ( 1) N 7 - B 10
( 76.47%) 0.8745* N 7 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.7807
-0.0082 -0.0679 -0.0137 0.0003 0.0000
0.0011 0.0000 0.0000 -0.0071 0.0085
( 23.53%) 0.4851* B 10 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.8251
-0.0456 0.0298 -0.0355 -0.0003 0.0000
0.0065 0.0000 0.0000 -0.0447 0.0206
8. (1.82090) BD ( 2) N 7 - B 10
( 88.21%) 0.9392* N 7 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0006 0.0000 1.0000 -0.0003
0.0000 -0.0028 -0.0037 0.0000 0.0000
( 11.79%) 0.3433* B 10 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0007 0.0000 0.9976 -0.0315
0.0000 0.0606 -0.0101 0.0000 0.0000
9. (1.98438) BD ( 1) N 7 - B 12
( 76.47%) 0.8745* N 7 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.4376
0.0080 -0.6501 0.0138 0.0006 0.0000
-0.0066 0.0000 0.0000 -0.0029 -0.0085
( 23.53%) 0.4851* B 12 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.4258
0.0090 0.7074 0.0571 -0.0006 0.0000
-0.0414 0.0000 0.0000 -0.0182 -0.0206
10. (1.98438) BD ( 1) N 8 - B 10
( 76.47%) 0.8745* N 8 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.3442
-0.0159 0.7041 0.0000 -0.0004 0.0000
0.0058 0.0000 0.0000 -0.0043 -0.0085
( 23.53%) 0.4851* B 10 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.3997
-0.0540 -0.7224 -0.0208 0.0004 0.0000
0.0365 0.0000 0.0000 -0.0267 -0.0206
11. (1.98438) BD ( 1) N 8 - B 11
( 76.47%) 0.8745* N 8 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 -0.3316
0.0159 0.7101 -0.0003 -0.0006 0.0000
0.0056 0.0000 0.0000 0.0045 0.0085
( 23.53%) 0.4851* B 11 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 0.3868
0.0536 -0.7294 -0.0217 0.0006 0.0000
0.0355 0.0000 0.0000 0.0280 0.0206
12. (1.82091) BD ( 2) N 8 - B 11
( 88.21%) 0.9392* N 8 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0007 0.0000 1.0000 -0.0003
0.0000 0.0046 -0.0005 0.0000 0.0000
( 11.79%) 0.3433* B 11 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0006 0.0000 0.9976 -0.0315
0.0000 -0.0215 0.0575 0.0000 -0.0001
13. (1.98438) BD ( 1) N 9 - B 11
( 76.47%) 0.8745* N 9 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 -0.6209 -0.0043 0.0001 0.7818
-0.0079 0.0540 0.0138 0.0002 0.0000
-0.0008 0.0000 0.0000 -0.0072 0.0085
( 23.53%) 0.4851* B 11 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
0.0003 -0.5587 0.0174 -0.0032 -0.8255
-0.0450 -0.0151 0.0363 -0.0002 0.0000
-0.0049 0.0000 0.0000 -0.0449 0.0206
14. (1.98438) BD ( 1) N 9 - B 12
( 76.47%) 0.8745* N 9 s( 38.55%)p 1.59( 61.44%)d 0.00( 0.01%)
0.0000 0.6209 0.0043 -0.0001 0.4492
0.0077 0.6422 -0.0139 -0.0003 0.0000
0.0067 0.0000 0.0000 -0.0026 -0.0085
( 23.53%) 0.4851* B 12 s( 31.25%)p 2.19( 68.50%)d 0.01( 0.25%)
-0.0003 0.5587 -0.0174 0.0032 -0.4383
0.0080 -0.6997 -0.0573 0.0003 0.0000
0.0420 0.0000 0.0000 -0.0168 -0.0206
15. (1.82092) BD ( 2) N 9 - B 12
( 88.21%) 0.9392* N 9 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
0.0000 0.0000 0.0000 0.0000 -0.0003
0.0000 0.0007 0.0000 1.0000 -0.0003
0.0000 -0.0018 0.0043 0.0000 0.0000
( 11.79%) 0.3433* B 12 s( 0.00%)p 1.00( 99.62%)d 0.00( 0.38%)
0.0000 0.0000 0.0000 0.0000 -0.0004
0.0000 0.0006 0.0000 0.9976 -0.0315
0.0000 -0.0391 -0.0474 0.0000 0.0000
16. (1.99943) CR ( 1) N 7 s(100.00%)p 0.00( 0.00%)
1.0000 0.0001 0.0000 0.0000 0.0001
0.0000 0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
Second order perturbation theory analysis of Fock Matrix in NBO basis
I only selected the ones whose E2 value is larger than 20kcal/mol.
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
within unit 1 8. BD ( 2) N 7 - B 10 /120. BD*( 2) N 9 - B 12 37.57 0.33 0.100 12. BD ( 2) N 8 - B 11 /113. BD*( 2) N 7 - B 10 37.57 0.33 0.100 15. BD ( 2) N 9 - B 12 /117. BD*( 2) N 8 - B 11 37.56 0.33 0.100
Natural bond orbital
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H6B3N3)
1. BD ( 1) H 1 - B 11 1.98670 -0.40393 115(v),119(v),56(v),66(v)
2. BD ( 1) H 2 - N 9 1.98495 -0.61481 114(v),116(v),119(g),118(g)
86(v),96(v)
3. BD ( 1) H 3 - B 12 1.98670 -0.40393 118(v),112(v),66(v),46(v)
4. BD ( 1) H 4 - N 7 1.98495 -0.61481 119(v),115(v),112(g),114(g)
76(v),96(v)
5. BD ( 1) H 5 - B 10 1.98670 -0.40393 116(v),114(v),46(v),56(v)
6. BD ( 1) H 6 - N 8 1.98495 -0.61482 112(v),118(v),116(g),115(g)
76(v),86(v)
7. BD ( 1) N 7 - B 10 1.98438 -0.68871 114(g),111(v),109(g),108(v)
97(v),119(v)
8. BD ( 2) N 7 - B 10 1.82090 -0.27139 120(v),102(v),98(v),35(v)
113(g)
9. BD ( 1) N 7 - B 12 1.98438 -0.68869 112(g),107(v),109(g),110(v)
77(v),115(v)
10. BD ( 1) N 8 - B 10 1.98438 -0.68870 116(g),109(v),111(g),106(v)
87(v),118(v)
11. BD ( 1) N 8 - B 11 1.98438 -0.68873 115(g),107(v),111(g),110(v)
77(v),112(v)
12. BD ( 2) N 8 - B 11 1.82091 -0.27140 113(v),82(v),78(v),43(v)
117(g)
13. BD ( 1) N 9 - B 11 1.98438 -0.68868 119(g),111(v),107(g),108(v)
97(v),114(v)
14. BD ( 1) N 9 - B 12 1.98438 -0.68872 118(g),109(v),107(g),106(v)
87(v),116(v)
15. BD ( 2) N 9 - B 12 1.82092 -0.27139 117(v),92(v),88(v),27(v)
120(g)
16. CR ( 1) N 7 1.99943 -14.13097 97(v),77(v),112(g),114(g)
17. CR ( 1) N 8 1.99943 -14.13097 87(v),77(v),116(g),115(g)
18. CR ( 1) N 9 1.99943 -14.13097 97(v),87(v),119(g),118(g)
19. CR ( 1) B 10 1.99917 -6.65247 114(v),116(v),109(v),111(v)
20. CR ( 1) B 11 1.99917 -6.65247 115(v),119(v),111(v),107(v)
21. CR ( 1) B 12 1.99917 -6.65247 118(v),112(v),107(v),109(v)
22. RY*( 1) H 1 0.00025 0.73512
IR spectrum

Charge distribution
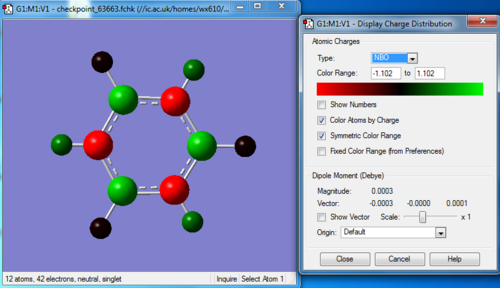
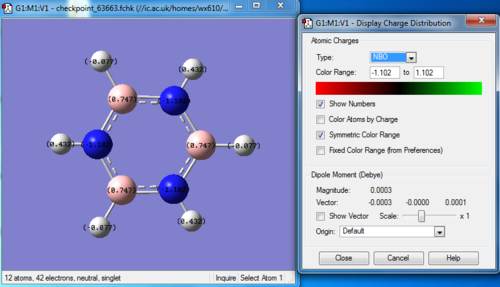
Bond information
| Bond Distance B-H | 1.19494 Å |
| Bond Distance N-H | 1.00974 Å |
| Average Bond Distance N-B | 1.43068 Å |
| Average Bond Angle H-B-N | 121.439 |
| Average Bond Angle H-N-B | 118.561 |
File link
Optimization:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20715
Frequency:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20717
NBO:DOI:10042/to-https://spectradspace.lib.imperial.ac.uk:8443/dspace/handle/10042/20718
Comparison
| Name | Charge distribution | Charge distribution by number |
|---|---|---|
| Benzene | 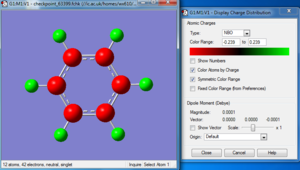 |
 |
| Boratabenzene | 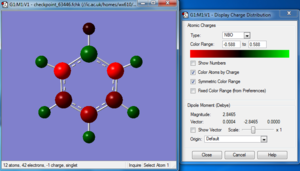 |
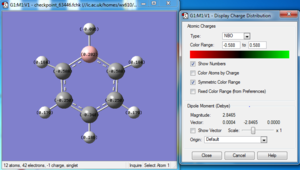 |
| Pyridinium | 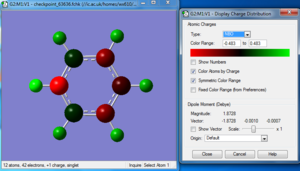 |
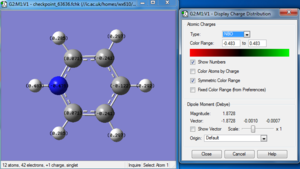 |
| Borazine | 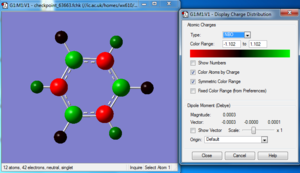 |
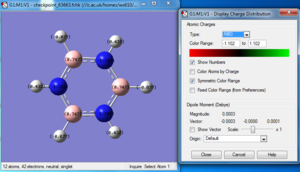 |
| MO energy level | Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|---|
| 17 (all pi bonding) | 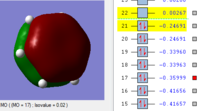 |
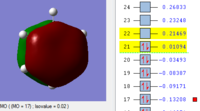 |
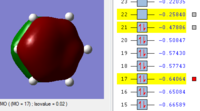 |
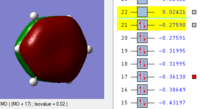 |
| HOMO | 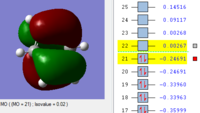 |
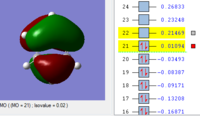 |
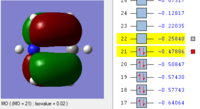 |
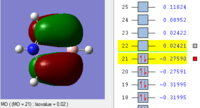 |
| LUMO | 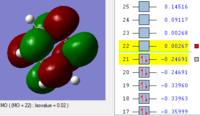 |
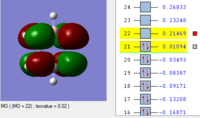 |
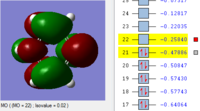 |
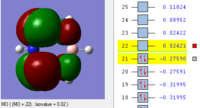 |
All have very similar totally bonding pi MO, with slightly outstretching electronegative atom for B atom and more for N atom. Pyridinium has the lowest energy and boratabenzene has the highest one for energy level 17. Benzene and borazine has similar energy.
For the HOMO, benzene and boratabenzene have similar MO shape while pyridinium and borazin share similar MO. The difference for the first pair is that B attracts more electrons, resulting relatively larger upper MO compared with benzene. Pyridinium has the lowest energy and boratabenzene has the highest one for HOMO. Benzene and borazine has similar energy.
As for the LUMO, all share similar MO. But N attracts more electrons and has larger MO towards itself. Pyridinium has the lowest energy and borazine has the highest one for LUMO. Boratabenzene is the second highest in energy and benzene is the second lowest in energy.