Rep:Mod:THISISMYDOMAIN272
NH3 molecule
NH3 molecule |
NH3 calculation results
Calculation method = RB3LYP
Basis set = 6-31G(d.p)
Final energy E(RB3LYP) = -56.55777 ±0.00001 a.u.
RMS gradient = 0.00000485 a.u.
Point group of molecule = C3V
N-H bond distance = 1.02±0.01Å
H-N-H bond angle = 106±1°
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986269D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
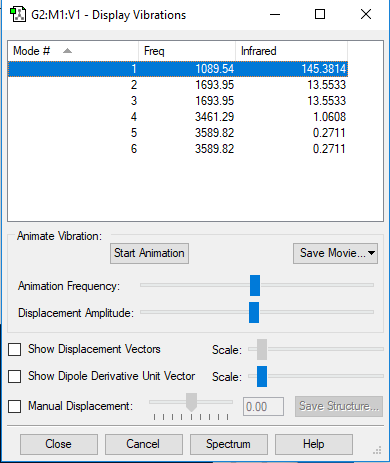
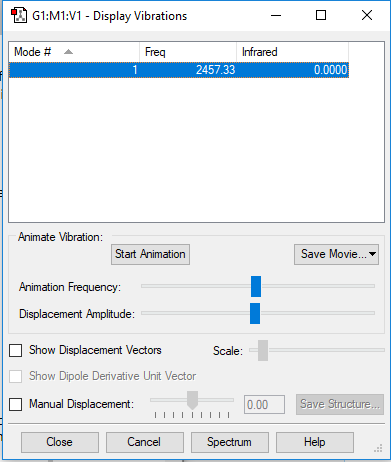
NH3 vibration results
NH3 vibration results Q and A
Based on the 3N-6 rule, as N=4 for NH3, 6 vibrational modes are expected. This matches the simulation with frequencies 2 and 3 and frequencies 5 and 6 being degenerate. Frequencies 1, 2 and 3 are bond bending modes whilst frequencies 4, 5 and 6 are bond stretching modes. Frequency 4 is highly symmetric, with frequency 1 seeming to be the umbrella mode. 2 bands would be expected to be seen in an experimental spectrum, being frequency 1 and an overlap of 2 and 3. This is because frequencies 2 and 3 are degenerate in energy so would produce a peak of equal intensity and wavenumber, appearing as a single band rather than 2.
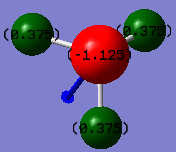
NH3 charge analysis
The Nitrogen and Hydrogen were calculated to have charges of -1.125 and 0.375 respectively. This matches what would be expected, as Nitrogen is more electronegative than Hydrogen, so would have a more negative charge. Each Hydrogen is in an identical environment so should each have equal charges, as depicted in the simulation.
The Gaussian optimisation file used is here
N2 molecule
N2 molecule |
N2 calculation results
Calculation method = RB3LYP
Basis set = 6-31G(d.p)
Final energy E(RB3LYP) = -109.52413±0.00001a.u.
RMS gradient = 0.00000060 a.u.
Point group of molecule = D∞h
N≡N bond distance = 1.11±0.01Å
N≡N bond angle = 180±1°
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401033D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
N2 vibration results
| Frequency 1 | |
|---|---|
| Wavenumber/cm-1 | 2457 |
| Symmetry | SGG |
| Intensity/a.u. | 0 |
| Image | 
|
As this is a linear molecule, the 3N-5 rule applies here. As there are 2 atoms in the molecule, N=2 so there should be 1 vibrational mode. This is equal to the number of calculated modes. Frequency 1 is a symmetric stretch that would not be expected to appear in an IR spectrum, but would in a Raman spectrum.
N2 charge analysis
Both Nitrogen's were calculated as being neutral, which is expected as they are equal in electronegativities so would be a non-polar molecule.
N2 Comparison to mono-metallic TM complex
In [CILSEV] from Conquest, the N≡N bonds are 1.092Å (N1≡N2), 1.124Å(N3≡N4) and 1.104Å (N5≡N6) respectively for mer-tris(Dinitrogen)-tris(di-n-propyl-phenylphosphine)-molybdenum whilst the calculated N≡N via B3LYP is 1.11Å. The N1≡N2 and N5≡N6 bonds are 0.019Å and 0.13Å shorter than the computational value due to the reduced orbital overlap between Mo and both N1 and N5. This is caused by the bulky phosphine groups which repel the 2 nitrogen ligands together and cause deviations in the expected bond angles and lengths of the complex. The N3≡N4 bond is 0.014Å larger than the calculated value, which can be attributed to the calculation having few iterations of relatively low resolution as well as the phophine group possibly lengthening the N3≡N4 bond. This can be improved by using a more accurate computational method which is more suited for mono-metallic complexes to get a better estimate for the bond length. The N3≡N4 bond is also expected to be slightly larger due to the transition metal withdrawing electron density from the N3≡N4 ligand, decreasing the N3≡N4 bond electron density and therefore increasing the bond length.
The Gaussian optimisation file for N2 is here
H2 molecule
H2 molecule |
H2 calculation results
Calculation method = RB3LYP
Basis set = 6-31G(d.p)
Final energy E(RB3LYP) = -1.17854±0.00001 a.u.
RMS gradient = 0.00000017 a.u.
Point group of molecule = D∞h
H-H bond distance = 0.74±0.01Å
H-H bond angle = 180±1°
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000005 0.001800 YES
RMS Displacement 0.000007 0.001200 YES
Predicted change in Energy=-1.939322D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
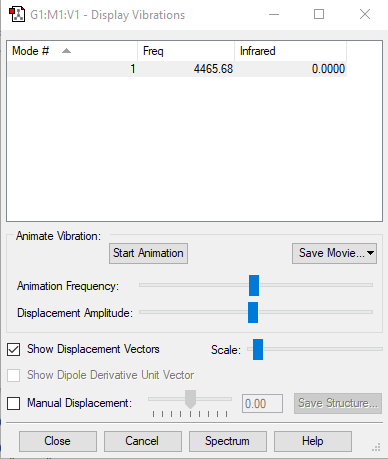
H2 vibration results
| Frequency 1 | |
|---|---|
| Wavenumber/cm-1 | 4466 |
| Symmetry | SGG |
| Intensity/a.u. | 0 |
| Image | 
|
As this is a linear molecule, the 3N-5 rule applies here. As there are 2 atoms in the molecule, N=2 so there should be 1 vibrational mode. This is equal to the number of calculated modes. Frequency 1 is a symmetric stretch that would not be expected to appear in an IR spectrum, but would in a Raman spectrum.
H2 charge analysis
Both Hydrogen's were calculated as being neutral, which is expected as they are equal in electronegativities so would be a non-polar molecule.
The Gaussian optimisation file for H2 is here
Haber-Bosch process energy calculation
E(NH3)= -56.55776873 a.u.
2*E(NH3)= -113.1155375 a.u.
E(N2)=-109.52412868 a.u.
E(H2)=-1.17853936 a.u.
3*E(H2)= -3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)] ≈ -0.05579 Hartree ≈ -146.5 kJmol-1
The ammonia product is more stable as it is lower in energy than the reagants, as shown in how the ΔE<0 which depicts an exothermic reaction.
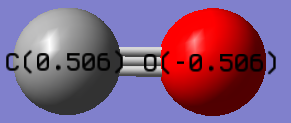
Project molecule, CO =
CO molecule |
CO calculation results
Calculation method = RB3LYP
Basis set = 6-31G(d.p)
Final energy E(RB3LYP) = -113.30945±0.00001 a.u.
RMS gradient = 0.00000433 a.u.
Point group of molecule = C ∞V
C≡O bond distance = 1.14±0.01Å
C≡O bond angle = 180±1°
Item Value Threshold Converged?
Maximum Force 0.000007 0.000450 YES
RMS Force 0.000007 0.000300 YES
Maximum Displacement 0.000003 0.001800 YES
RMS Displacement 0.000004 0.001200 YES
Predicted change in Energy=-2.221214D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1379 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
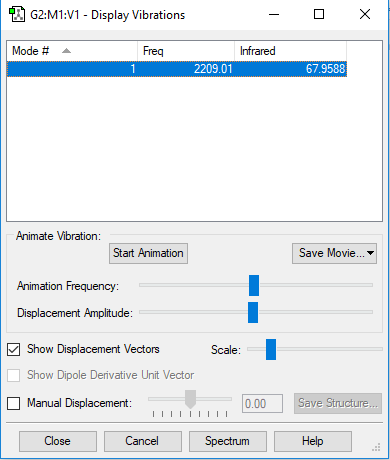
CO vibration results
| Frequency 1 | |
|---|---|
| Wavenumber/cm-1 | 2209 |
| Symmetry | SG |
| Intensity/a.u. | 68 |
| Image | 
|
As this is a linear molecule, the 3N-5 rule applies here. As there are 2 atoms in the molecule, N=2 so there should be 1 vibrational mode. This is equal to the number of calculated modes. Frequency 1 is a symmetric stretch that would not be expected to appear in an IR spectrum, but would in a Raman spectrum.
CO charge analysis
The Carbon and Oxygen were calculated to have charges of 0.506 and -0.506 respectively. This is as predicted as Oxygen is more electronegative than Carbon, so would be expected to be more negatively charged than Carbon. This would also imply that they should have equal magnitudes of charge as they are the only 2 atoms in the molecule.
The Gaussian optimisation file used is here
CO molecular Orbital analysis
MOs 5 and 6 are 2 degenerate MOs as they are the overlap of different p AO but at different orientations, being in the x and then the y plane. MO 7 is the highest occupied molecular orbital and would act as a nucleophile from the Carbon as it has a greater charge density than the Oxygen. MO 8 is the lowest unoccupied molecular orbital with the carbon having a more diffuse spread of electron density, so would be more likely to be attacked by a nucleophile. MO's 7 and 8 are antibonding orbitals whilst MO's 4, 5 and 6 are bonding orbitals.
Independence mark, calculating the N2 bond length via a different computational method
N2 molecule |
N2 calculation results
Calculation method = RCAM-B3LYP
Basis set = 6-31G(d.p)
Final energy E(RCAM-B3LYP) = -109.47916±0.00001 a.u.
RMS gradient = 0.00000012 a.u.
Point group of molecule = D∞V
C≡O bond distance = 1.10±0.01Å
C≡O bond angle = 180±1°
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-1.203893D-14
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1008 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
N2 vibration results
| Frequency 1 | |
|---|---|
| Wavenumber/cm-1 | 2511 |
| Symmetry | SGG |
| Intensity/a.u. | 0 |
| Image | 
|
This is a linear molecule, so the 3N-5 rule applies here. As there are 2 atoms in the molecule, N=2 so there should be 1 vibrational mode. This is equal to the number of calculated modes. Frequency 1 is a symmetric stretch that would not be expected to appear in an IR spectrum, but would in a Raman spectrum.
N2 charge analysis
Both Nitrogen's were calculated as being neutral with CAM-B3LYP, which is expected as they are equal in electronegativities so would be a non-polar molecule.
Comparing B3LYP and CAM-B3LYP results for N2
Comparing the 2 vibrational values, B3LYP=2457cm-1 whilst RCAM-B3LYP=2511cm-1. The experimental value for N2 is 2330cm-1 as shown here. This shows that B3LYP and CAM-B3LYP have percentage uncertainties of 5.3% and 7.5% respectively. This suggests that B3LYP is more accurate at calculating frequencies than CAM-B3LYP.
Comparing the bond lengths, B3LYP=1.11Å whilst RCAM-B3LYP=1.10Å. The experimental value for N2 is 1.0975Å[1]]. This shows that B3LYP and CAM-B3LYP have percentage uncertainties of 0.11% and 0.023% respectively. This shows that CAM-B3LYP is more accurate than B3LYP when calculating bond lengths.
Both charge distributions calculated were the same, being 0 on each Nitrogen.
Using the N2 bond length from CAM-B3LYP to compare with the monometallic complex
The CAM-B3LYP length of 1.10Å compared to the 3 N≡N of 1.092Å (N1≡N2), 1.124Å(N3≡N4) and 1.104Å (N5≡N6) from [CILSEV] shows that the calculated value is now also shorter than N5≡N6. This is likely due to how, whilst N5≡N6 and N1≡N2 are shortened by the large phoshine groups, the effect of the transition metal withdrawing electron density from the N≡N is greater. This causes each N≡N in the monometallic structure to be longer than expected. This is different to the conclusion made via the B3LYP calculation as the bond length of 1.11Å was larger than both N1≡N2 and N5≡N6, suggesting that the shortening by the large phosphine groups was greater than the lengthening cased by the transition metal.
The Gaussian optimisation file used is here
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES, However you have given a bond angle of 180 for N2 and H2, there are no bond angles in diatomic molecules. Bond angles involve exactly 3 atoms.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4/5
Have you completed the calculation and included all relevant information?
YES, However you have given a bond angle of 180, there are no bond angles in diatomic molecules. Bond angles involve exactly 3 atoms.
Have you added information about MOs and charges on atoms?
YES, overall good explanations, nicely laid out, well done! You could have added a bit more detail on the MOs for example why does the C contribute more strongly to the HOMO and LUMO?
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far
You did an extra calculation on N2 and looked at a further literature comparison of the bond length well done!