Rep:Mod:SOL0951
Ammonia
Optimisation
Optimisation was carried out using Guassview 5.0. The optimisation file can be found here.
NH3 Model |
| Molecule | NH3 (ammonia) |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy (a.u.) | -56.55776873 |
| RMS Gradient Norm (a.u.) | 0.00000485 |
| Point Group | C3V |
Optimised N-H bond distance: 1.01789 Å
Optimised H-N-H bond angle: 105.741°
The values generated are very comparable to experimentally determined literature values. Most sources give the N-H bond angle as 106.7° and the bond legnth as 1.018 Å [1]
The item table created by Gaussian, provided as proof that the optimization algorithm has converged, can be found below:
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986274D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
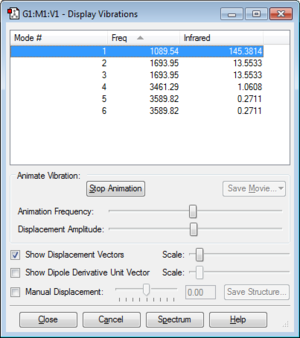
Vibrations and Charges
6 modes are expected form the 3N-6 rule. Modes 2 and 3, and modes 5 and 6, are degenerate. 1, 2 and 3 are bending vibrations. 4,5 and 6 are stretching vibrations. Mode 1 is a symetric bend. Mode 4 is a symetric stretch. Mode 1 is known as the "umbrella" mode. 4 bands would be expecte in an experimental spectrum of gaseous ammonia.
Calculated charges on the different atoms in this molecule:
N-atom: -1.125 e
H-atom: +0.375 e
We expect the highly electronegative nitrogen to withdraw electrons from the hydrogens, making it more negative and leaving the hydrogens more positive. The charges given by Gaussian agree with our predictions.
Reaction Energy
N2
The optimisation file can be found here.
N2 Model |
| Molecule | N2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy (a.u.) | -109.52412868 |
| RMS Gradient Norm (a.u.) | 0.00000060 |
| Point Group | D*H |
Optimised N≡N bond distance: 1.10550 Å
Proof that the optimization algorithm has converged can be found below:
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400948D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
| Mode | Frequency | Infrared |
|---|---|---|
| 1 | 2457.33 | 0.0000 |
H2
The optimisation file can be found here.
H2 Model |
| Molecule | H2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy (a.u.) | -1.17853936 |
| RMS Gradient Norm (a.u.) | 0.00000017 |
| Point Group | D*H |
Optimised H-H bond distance: 0.74279 Å
Proof that the optimization algorithm has converged can be found below:
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
| Mode | Frequency | Infrared |
|---|---|---|
| 1 | 4465.68 | 0.0000 |
Energy for the reaction of N2 + 3H2 -> 2NH
The energy change for this reaction can be calculated by using the energy values generated by Gaussview.
E(NH3)= -56.55776873 Eh
2*E(NH3)= -113.1155375 Eh
E(N2)= -109.52412868 Eh
E(H2)= -1.17853936 Eh
3*E(H2)= -3.53561808 Eh
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579074 Eh
ΔE= -146.48 kJ/mol
The energy change when converting hydrogen and nitrogen gas into ammonia gas is negative, so the ammonia product is more stable than the gaseous reactants.
The Cyanide Ion
The optimisation file can be found here.
CN Model |
| Molecule | CN- |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy (a.u.) | -92.82453153 |
| RMS Gradient Norm (a.u.) | 0.00000704 |
| Point Group | C*v |
| Mode | Frequency | Infrared |
|---|---|---|
| 1 | 2139.19 | 7.7521 |
| Atom | Charge (e) |
|---|---|
| Carbon | -0.584 |
| Nitrogen | -0.416 |
Optimised C=N bond length of 1.18408 Å. This is close to literature values, which cite the bond angle as being 1.16 Å [2].
Proof that the optimization algorithm has converged can be found below:
Item Value Threshold Converged?
Maximum Force 0.000012 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000005 0.001800 YES
RMS Displacement 0.000008 0.001200 YES
Predicted change in Energy=-6.650396D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1841 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Molecular Orbitals
- ↑ Haynes, William M., ed. (2013). CRC Handbook of Chemistry and Physics (94th ed.). CRC Press. pp. 9–26. ISBN 9781466571143.
- ↑ http://hydra.vcp.monash.edu.au/modules/mod2/bondlen.html.