Rep:Mod:RA8410
Module 1
Part 1
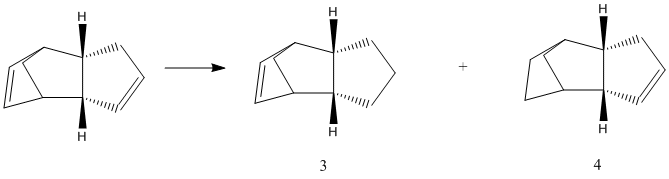
Cyclopentadiene Dimerisation
The dimerisation of cyclopentadiene is a common example of a Diels-Alder reaction, a 4+2 cycloaddition. It is a thermally activated reaction, with 6 pi electrons in the transition state, and leads to the necessity of storing cyclopentadiene at low temperature and 'cracking' before using it in a reaction.
Although stereochemical relationships are preserved, e.g. cis dienophiles have a syn relationship in the product, there are 2 possible isomers produced as a result of 2 different transition states. An exo trnaisition state (producing isomer 1) involves overlap of reaction centres, ie the non-reacting parts of the molecules are staggered. The endo transition state is normally preferred as a result of favourable interactions that occur when the molecules completely overlap, i.e. are eclipsed(isomer 2).
It is observed that isomer 2 is less stable than isomer 1. This, along with the fact that the endo preference is a result of forces in the transition state, suggests the reaction is under kinetic control and that the preferred isomer is in fact the less thermodyamically favourable isomer. Reasons for this are provided by analysis of the data provided in the minimisation. A high positive value means destabilisation relative to normal values. The major difference between 1 and 2 arises as a result of torsional strain. There appears to be more in isomer 2, causing a significant difference of 8.8 kJ/mol.
1
------------MM2 Minimization------------ Note: All parameters used are finalized (Quality = 4). Iteration 130: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 1.2848 Bend: 20.5806 Stretch-Bend: -0.8382 Torsion: 7.6550 Non-1,4 VDW: -1.4168 1,4 VDW: 4.2336 Dipole/Dipole: 0.3775 Total Energy: 31.9 kcal/mol Calculation completed ------------------------------------
2
------------MM2 Minimization------------ Note: All parameters used are finalized (Quality = 4). Iteration 142: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 1.2507 Bend: 20.8487 Stretch-Bend: -0.8358 Torsion: 9.5103 Non-1,4 VDW: -1.5442 1,4 VDW: 4.3202 Dipole/Dipole: 0.4476 Total Energy: 34.0 kcal/mol Calculation completed ------------------------------------
Hydrogenation
After the dimer is formed, its hydrogenation is selective for one position. There are regioselectivity issues possible because of the 2 double bonds, which begs the question which is the preferred position. Analysis shows that 4 is in fact the preferred regioisomer. In fact, the energies differ by around 18.8 kJmol-1. There is even around 3 kJmol-1 stabilisation relative to the thermodynamically more stable dimerisation product. It is therefore possible to estimate the energy of hydrogenation in both positions. Isomer 4 has a reaction free energy change of -11.7 kJmol-1 while isomer 3 has a free energy change of +7.1kJmol-1. The different parameters from the MM2 calculation allow comparison of the structures in order to rationalise the disparity in energy of the compounds. It should first be noted that the stretching energy appears seems to be very similar in all cases. The largest difference comes as a result of bending. This is likely due to decreased bending from eq. geometries. This is something which has a lower value in both products than in both reactants. The results for torsion actually show greater torsional strain in 4 despite its higher stability. This suggests that 4 is more likely to encounter torsional strain. This is possibly due to the location of the new hydrogen atoms. In 4, they are closer to the axial carbon, giving a higher chance of 1,3 diaxial interactions, in isomer 3, these hydrogens are pointing away from the system due to the sp2 system. The magnitude of energy change as a result of dipole/dipole interactions is significantly lower in both products. One possible explanation is the less sterically congested structure. It follows that there is less distortion in product 4, which suggests that the hydrogenated position is the alkene which gives more bending from equilibrium
The distortion from ideal bond lengths and angles in such a system makes it easier for the molecule to possess a dipole, something less likely to happen in the hydrogenated products.
3
------------MM2 Minimization------------ Note: All parameters used are finalized (Quality = 4). Iteration 109: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 1.2776 Bend: 19.8561 Stretch-Bend: -0.8349 Torsion: 10.8116 Non-1,4 VDW: -1.2204 1,4 VDW: 5.6328 Dipole/Dipole: 0.1621 Total Energy: 35.7 kcal/mol Calculation completed ------------------------------------
4
------------MM2 Minimization------------ Note: All parameters used are finalized (Quality = 4). Iteration 134: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 1.0976 Bend: 14.5247 Stretch-Bend: -0.5495 Torsion: 12.4975 Non-1,4 VDW: -1.0702 1,4 VDW: 4.5114 Dipole/Dipole: 0.1406 Total Energy: 31.2 kcal/mol Calculation completed ------------------------------------
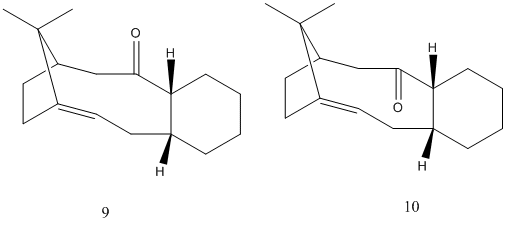
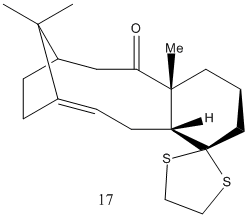
Taxol Synthesis Intermediate
In defiance of Bredt's rule, some alkenes at bridgeheads have high stabilities. This is likely to occur in medium size polycyclic alkenes(DOI:10.1021/ja00398a003). This intermediate is, of course, bicyclic, consisting of a 6 membered ring and a 10 membered ring. The position of the alkene imparts extra stability, making hydrogenation unlikely and further reaction in general less favoured. Interestingly, the values produced by MM2 and MMFF94 treatments lead to radically different values. It is important to note, however, that the MMFF94 optimisation still maintains a similar difference in energy between the 2 atropisomers. Therefore, as expected, relative energies of different configurations are possible, but comparison of energies between differently calculated values is not accurate. Comparison of the optimised structures shows significant differences. These differences certainly correlate with the energy difference, although there is not necessarily a causal link. It should be noted, for example, that in 9, the more stable isomer, the 6 member ring is in a more stable chair form, while 10 is in a conformation that more closely resembles a twist-boat. This may explain the greater amount of unfavourable 1,4 vdW forces and torsion. Both 10 member rings have a chair-like conformation, but differ from a chair due to the alkene. Similarly, kinetic instability is likely in 10, which has hydrogens antiperiplanar to the ketone. This means enolisation and reaction as a base is possible, something much less likely in 9 due to the lack of an antiperiplanar relationship. 9 does, however, possess an anti-periplanar relationship to a C-C bond, allowing donation into its antibonding orbital, weakly stabilising the molecule. The 6 membered ring seems to be the main difference. The down-pointing carbonyl group appears to alter the relative position of the ring atoms, giving a more stable configuration. It also appears to give rise to less eclipsed hydrogen atoms.
9
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 247: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.7852 Bend: 16.5393 Stretch-Bend: 0.4299 Torsion: 18.2542 Non-1,4 VDW: -1.5545 1,4 VDW: 13.1102 Dipole/Dipole: -1.7249 Total Energy: 47.8396 kcal/mol Calculation completed ------------------------------------
------------MMFF94 Minimization------------ Iteration 61: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Final Energy: 70.5333 kcal/mol Calculation completed -------------------------------------
10
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 646: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 3.1448 Bend: 19.7772 Stretch-Bend: 0.3069 Torsion: 21.6828 Non-1,4 VDW: -0.3041 1,4 VDW: 15.4212 Dipole/Dipole: -1.6934 Total Energy: 58.3354 kcal/mol Calculation completed ------------------------------------
------------MMFF94 Minimization------------ Iteration 64: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Final Energy: 82.3764 kcal/mol Calculation completed ------------------------------------
Regioselective Addition of Dichlorocarbene to a diene
Calculations were performed using the standard MM2 calculation and the more applicable MOPAC. Interestingly, the bond lengths matched up almost completely, with only the relative orientation of the atoms seemingly affected. The MOPAC calculation does, however, give an energy of 22 kcal/mol for heat of formation. This is much higher than the MM2 example suggesting that the re-optimised structure is actually less stable. This could be explained by greater accuracy.
Comparison of MM2 and MOPAC structures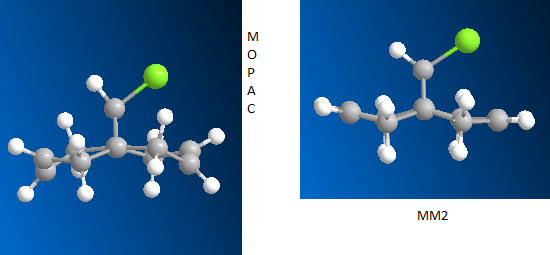
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 146: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 0.6187 Bend: 4.7384 Stretch-Bend: 0.0400 Torsion: 7.6590 Non-1,4 VDW: -1.0680 1,4 VDW: 5.7941 Dipole/Dipole: 0.1123 Total Energy: 17.8945 kcal/mol Calculation completed ------------------------------------
------------ Mopac Interface ------------ Model: Untitled-2 Mopac Job: AUX RM1 CHARGE=0 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.08889 (< 0.10000) Heat of Formation = 22.82771 Kcal/Mol -----------------------------------------
Gaussian MOs
MOs can then by calculated using the gaussian interface, ie DFT. These are useful because it shows the delocalisation of charge and the HOMO can be used to show which alkene is more likely to react with dichlorocarbene. Shape and size, i.e. the magnitude of the electron density, need to be considered as well as how the part nearest the alkene interacts with nearby lobes. The MOSs from LUMO+2 to HOMO-1 are shown below in order of descending energy, as they would appear in an MO diagram. Of course, as this is a nucleophilic compound, it is likely to react by donation of electrons, therefore the HOMO is particularly significant. Although the HOMO has a similar distribution of electron density in both alkenes, the MO is not symmetrical. As can be seen below, the presence of the chlorine atom affects the reactivity. Stabilising interactions can occur between the in phase nearby chlorine atom with part of the lobe near the alkene. This and similar interactions are observed to a greater extent, and the cumulative impact of these interactions means that the alkene on the opposite side to the chlorine atom is more reactive.
------------ Gaussian Interface ------------ Model: 12.mol 1) Gaussian Job: # B3LYP/6-31G Test Charges (Electron Density): C(1) 5.573431 C(2) 5.599455 C(3) 5.105279 C(4) 5.146899 C(5) 5.100133 C(6) 5.222397 C(7) 5.122766 C(8) 5.146250 C(9) 5.090452 C(10) 5.196826 C(11) 6.201393 Cl(12) 17.009635 H(13) 0.597370 H(14) 0.636511 H(15) 0.604224 H(16) 0.616749 H(17) 0.631204 H(18) 0.579557 H(19) 0.594687 H(20) 0.636964 H(21) 0.595531 H(22) 0.609023 H(23) 0.624058 H(24) 0.586562 H(25) 0.532917 Charges (Mulliken Charges): C(1) 0.058822 C(2) 0.077064 C(3) -0.268579 C(4) -0.144977 C(5) -0.086025 C(6) -0.339965 C(7) -0.259290 C(8) -0.145136 C(9) -0.100106 C(10) -0.305748 C(11) -0.433698 Cl(12) 0.050311 H(13) 0.154954 H(14) 0.128035 H(15) 0.131669 H(16) 0.124466 H(17) 0.117854 H(18) 0.185770 H(19) 0.153014 H(20) 0.130861 H(21) 0.136138 H(22) 0.132605 H(23) 0.127091 H(24) 0.166252 H(25) 0.208619 Gaussian Interface: Dipole = (1.8637, -0.0253, -0.7711) 2.0171 Debye Gaussian Interface: SCF Energy = -556464.69 Kcal/Mol --------------------------------------------
Charge Distribution
The charge distribution presents a similar trend to the data from the MOs. As noted in the gaussian output in ChemBio 3D shown above, only C1 and C2 have positive Mulliken charges. It can be seen here that although there is a very slight localisation of positive charge in the alkene facing away from Cl, there is a much larger positive charge density at the other alkene. This means that it is more likely to be electrophilic, rather than nucleophilic, which suggests that the alkene on the opposite side of the molecule to Cl is more nucleophilic and, as such, is more likely to be the site of reaction with an electrophile.
Frequencies
The frequencies were then calculated and an excerpt from the log file shows that the optimisation is likely to be complete. It was also used to model the molecules vibrations with a view to determining the difference between the two alkene environments.
Low frequencies --- -5.5284 -0.0020 -0.0018 -0.0008 7.6985 10.6459
Low frequencies --- 90.3938 125.6424 158.0780
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 90.3715 125.6349 158.0737
Red. masses -- 2.6009 2.6562 3.1947
Frc consts -- 0.0125 0.0247 0.0470
IR Inten -- 0.2023 0.3008 0.1998
The 2 C-Cl bond stretches are given below. The lower vibrational frequency value is less symmetrical in its effect on the molecule(particularly with regards to the cyclohexene rings) than the other, and this may explain its higher intensity, given that an IR active vibration is one which changes the overall dipole of the molecule.
| Frequency | Intensity |
|---|---|
| 770.96 | 25.1074 |
| 930.15 | 7.2775 |
Stretching frequency of the C=C is clearly an important factor in this reaction, this much is clear from the formation of product, which will see the C=C stretch as it becomes a carbon-carbon single bond, in other words this degree of freedom is likely to very significant in the formation of a transition state. This suggests, in contrast to the results above, that the Exo alkene actually is kinetically favoured as a consequence of its lower energy transition state formation.
| Frequency | Intensity | |
|---|---|---|
| Exo to Cl | 1736.99 | 4.1948 |
| Endo to Cl | 1757.30 | 3.9359 |
Monosaccharide chemistry and the mechanism of glycosidation
Sugar chemistry is famously complicated by the stereochemistry of the carbons in a sugar or the regioselectivity of any reactions, as there are generally many hydroxyls. As such, it is important that any reaction proceeds with known stereo- and regio-chemical outcome. The anomeric centre, in other words the site of the aldehyde in an open chain form, can exhibit either α(axial) or β(equatorial) comformation, and these properties are important to keep track of. In order to selectively product 1,2 trans products between the anomeric centre and C2, the use of a neighbouring group effect may be invoked. This is seen in this case, where after elimination of an anomeric leaving group by C5 oxygen, the C2 acetyl group forms an oxenium, leading to a 5 member ring. A nucleophile then attacks in an Sn2 type mechanism, leading, as mentioned, to a 1,2 trans product. Assuming constant stereochemistry at every carbon except the anomeric centre and C2, there are a total of 8 permutations, 4 for each intermediate in this reaction.
Intermediate A
Intermediate A has 4 isomers. These are shown below and result from axial or equatorial oreiantation of the acetyl group and the different positions of the carbonyl and methyl groups. The disctinction for the latter is more comformational, and these compounds will probably have an energy difference due to different steric and electronic properties of O and Me. Indeed, it is found that this is the case. Using the more optimised MOPAC energies, it is seen that there is a significant difference between 1 and 2, as well as between 3 and 4.
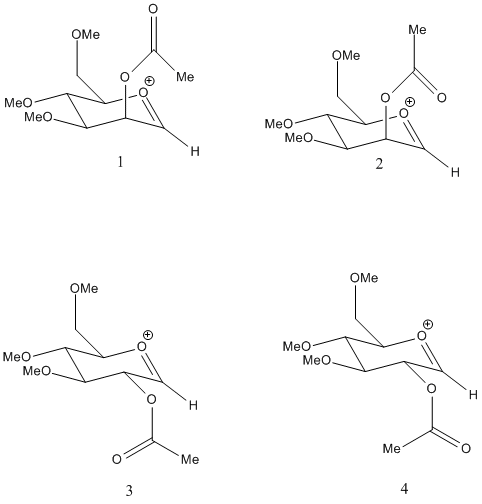
However, problems were encountered, as the ring appeared to undergo a ring flip during optimisation, interconverting axial and equatorial substituents. The experiment was repeated, but gave similar errors. This means that in this case the information is not particularly useful, but does lead to some observations useful in analysing the reaction.
It seems most useful to compare the 2 sets of stereoisomers, which are unlikely to interconvert. In each case, the orientation of the acetyl group means that there is a more reactive(but not necessarily stable) isomer, ie, the one where the carbonyl oxygen is oriented closer to the anomeric centre. Isomer 2 is expected to be the reactive conformation due to the orientation of the oxygen atom. This small change in conformation actually leads to an energy difference of around 10 kcal/mol. This seems to largely due to sterics, with the structure of 1 more bent from equilibrium. The opposite of the expected dipole/charge interactions is observed, with the interaction in 1 actually stabilising the molecule more. Isomer 3 is the most stable conformation, while isomer 4(by MM2) is the least stable, although in MOPAC the order changes somewhat. Nevertheless, an energy difference of around 20kcal/mol was observed. Surprisingly, the position of the oxygen atom actually led to a destabilising effect from charge/dipole interactions. This, in fact accounts for most of the difference, going from strongly stabilising in 3 to strongly destabilising in 4. As conversion of enantiomers is unlikely, 2 conformations for each enantiomer become clear, along with 2 contrasting patterns in terms of energy. In 1 and 2, 1 is the more unstable and also less reactive isomer. This means that the reaction will normally proceed via 2, which is likely to be the dominant conformation, giving a diaxial cyclic oxenium. In 3 and 4, 3 is far more stable, and as such may act as a kinetic trap, preventing conversion to 4, and hence limiting the reaction, which would produce a di-equatorial oxonium.
Isomer 1
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 918: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.4662 Bend: 14.4183 Stretch-Bend: 0.9592 Torsion: 2.2873 Non-1,4 VDW: -0.8958 1,4 VDW: 18.9786 Charge/Dipole: -13.2527 Dipole/Dipole: 7.8734 Total Energy: 32.8345 kcal/mol Calculation completed ------------------------------------
------------ Mopac Interface ------------ Model: sac1 Mopac Job: AUX AM1 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.06370 (< 0.10000) Heat of Formation = -64.04465 Kcal/Mol -----------------------------------------
Isomer 2
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 508: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.3143 Bend: 9.9856 Stretch-Bend: 0.8660 Torsion: 0.2886 Non-1,4 VDW: -2.6735 1,4 VDW: 18.6082 Charge/Dipole: -4.3853 Dipole/Dipole: 4.1430 Total Energy: 29.1468 kcal/mol Calculation completed ------------------------------------
Model: sac2.mol Mopac Job: AUX PM6 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.09062 (< 0.10000) Heat of Formation = -75.84911 Kcal/Mol -----------------------------------------
Isomer 3
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 888: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.2387 Bend: 10.2565 Stretch-Bend: 0.8836 Torsion: 2.7433 Non-1,4 VDW: -2.0317 1,4 VDW: 19.1268 Charge/Dipole: -12.0536 Dipole/Dipole: 5.3097 Total Energy: 26.4733 kcal/mol Calculation completed ------------------------------------
------------ Mopac Interface ------------ Model: sac3.mol Mopac Job: AUX PM6 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.09373 (< 0.10000) Heat of Formation = -85.01346 Kcal/Mol -----------------------------------------
Isomer 4
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 644: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.5414 Bend: 13.1650 Stretch-Bend: 1.0871 Torsion: 2.1793 Non-1,4 VDW: -3.0066 1,4 VDW: 18.4322 Charge/Dipole: 8.8154 Dipole/Dipole: 4.5049 Total Energy: 47.7187 kcal/mol Calculation completed ------------------------------------
------------ Mopac Interface ------------ Model: sac4.mol Mopac Job: AUX PM6 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.08623 (< 0.10000) Heat of Formation = -67.69883 Kcal/Mol -----------------------------------------
Oxenium cations
The oxenium intermediate can exist in 4 different forms. These are diaxial, diequatorial, axial-equatorial, and equatorial-axial). These can be considered similarly to cis and trans. It is known that the attack of the nucleophile occurs with inversion of stereochemistry, ie a mechanism similar to Sn2, and produces 1,2 trans products. This suggests that only the cis oxeniums are involved in the reaction. This is seen in 1 and 2 which are more stable than 3 or 4. Naturally, a source of difference is the strained nature of the 5 membered ring when forced into a trans-like configuration. Similarly, a much higher bend value means that the molecules are more bent out of their natural geometries. Therefore, in addition to the weight of evidence from these calculations, as well as the observations of many carbohydrate chemists, the trans-like isomers are much higher in energy and therefore less likely to exist in any appreciable amount. This means that the product is formed exclusively through 1 and 2. The relationship between the energies in MM2 and MOPAC is mainly preserved, although 3 and 4 are viewed as more similar in energy than would be expected.
1
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 616: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.0715 Bend: 13.7880 Stretch-Bend: 0.6455 Torsion: 7.4761 Non-1,4 VDW: -2.8174 1,4 VDW: 18.4525 Charge/Dipole: -10.5251 Dipole/Dipole: 0.7207 Total Energy: 29.8118 kcal/mol Calculation completed ------------------------------------
------------ Mopac Interface ------------ Model: monoint1.mol Mopac Job: AUX PM6 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.08705 (< 0.10000) Heat of Formation = -87.10934 Kcal/Mol -----------------------------------------
2
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 722: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.0760 Bend: 13.9558 Stretch-Bend: 0.7325 Torsion: 7.0901 Non-1,4 VDW: -2.5143 1,4 VDW: 17.4912 Charge/Dipole: -10.5192 Dipole/Dipole: -0.5518 Total Energy: 27.7603 kcal/mol Calculation completed ------------------------------------
------------ Mopac Interface ------------ Model: monoint2.mol Mopac Job: AUX PM6 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.09322 (< 0.10000) Heat of Formation = -91.66280 Kcal/Mol -----------------------------------------
3
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 454: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.7557 Bend: 17.9996 Stretch-Bend: 0.8567 Torsion: 7.9814 Non-1,4 VDW: -2.2914 1,4 VDW: 19.1755 Charge/Dipole: -1.0318 Dipole/Dipole: -0.1213 Total Energy: 45.3243 kcal/mol Calculation completed ------------------------------------
------------ Mopac Interface ------------ Model: monoint3.mol Mopac Job: AUX PM6 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.09702 (< 0.10000) Heat of Formation = -66.86476 Kcal/Mol -----------------------------------------
4
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 505: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.7128 Bend: 17.4577 Stretch-Bend: 0.7962 Torsion: 8.1734 Non-1,4 VDW: -2.4523 1,4 VDW: 19.4013 Charge/Dipole: -0.3166 Dipole/Dipole: -1.7463 Total Energy: 44.0263 kcal/mol Calculation completed ------------------------------------
------------ Mopac Interface ------------ Model: monoint4mopac.mol Mopac Job: AUX PM6 CHARGE=1 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.09524 (< 0.10000) Heat of Formation = -66.85511 Kcal/Mol -----------------------------------------
Conclusion
It is clear that 1,2 cis is required in the oxenium ion intermediate and this will lead to a 1,2 trans product. It is also shown that these are produced from 2 and 4, ie axial and equatorial acetyl groups with the Oxygen atom pointing towards the anomeric centre. Both, of course, produce 1,2 trans, but in 2 will produce the α anomer and 4 will produce the β anomer.
Part 2
Taxol Intermediate
Optimisation
Optimised Conformation of intermediate
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 559: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 5.3610 Bend: 20.7602 Stretch-Bend: 0.7644 Torsion: 27.4493 Non-1,4 VDW: -0.5898 1,4 VDW: 17.4411 Dipole/Dipole: -2.4568 Total Energy: 68.7294 kcal/mol Calculation completed ------------------------------------
Item Value Threshold Converged?
Maximum Force 0.000029 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.001120 0.001800 YES
RMS Displacement 0.000192 0.001200 YES
Predicted change in Energy=-1.880521D-08
Optimization completed.
-- Stationary point found.
NMR
NMR spectra were then calculated using mpw1pw91/6-31G(d,p) calculations. Looking over the data, it would be easy to conclude that the downfield results of carbon-13 NMR are more accurate and that these overall are more accurate than proton NMR. This is not the case however, but rather this is because all results have the same error. This means that error will be more significant in smaller values. DOI:10042/23282
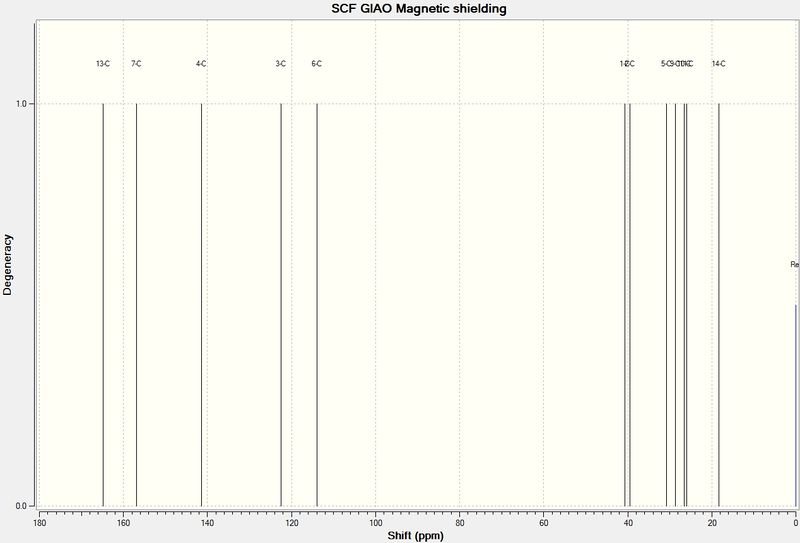
13C
A picture of the predicted NMR spectrum is shown below. There is also a table comparing the NMR data obtained from prediction and experiment. Resolution of both spectra is less thaqn desired, however, and the literature spectrum appears to have more carbon environments, suggesting impurities. Secondly, although the data could be viewed as a .txt file, allowing it to be tabulated, the overall resolution, especially in the lower, more alkyl dominated regions.
It is of little surprise that C-11 is in fact the ketone carbon and C-21, for example is a methyl group.
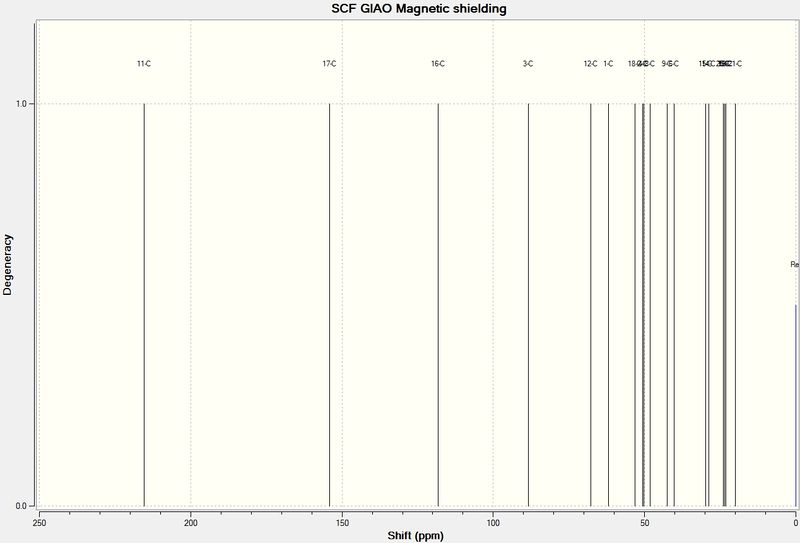
| Carbon | Predicted Chemical Shift (ppm) | Literature Chemical Shift (ppm) |
|---|---|---|
| 11-C | 215.5 | 218.79 |
| 17-C | 154.1 | 144.46 |
| 16-C | 118.3 | 125.33 |
| 3-C | 88.5 | 78.88 |
| 12-C | 67.7 | 56.19 |
| 1-C | 62.0 | 52.52 |
| 18-C | 53.2 | 48.50 |
| 2-C | 50.7 | 46.80 |
| 4-C | 50.2 | 45.76 |
| 8-C | 48.2 | 39.80 |
| 9-C | 42.5 | 38.91 |
| 6-C | 40.3 | 35.85 |
| 15-C | 29.8 | 28.79 |
| 14-C | 28.8 | 28.29 |
| 20-C | 24.0 | 26.88 |
| 5-C | 23.6 | 25.66 |
| 19-C | 23.6 | 23.86 |
| 13-C | 23.1 | 20.96 |
| 21-C | 20.1 | 18.71 |
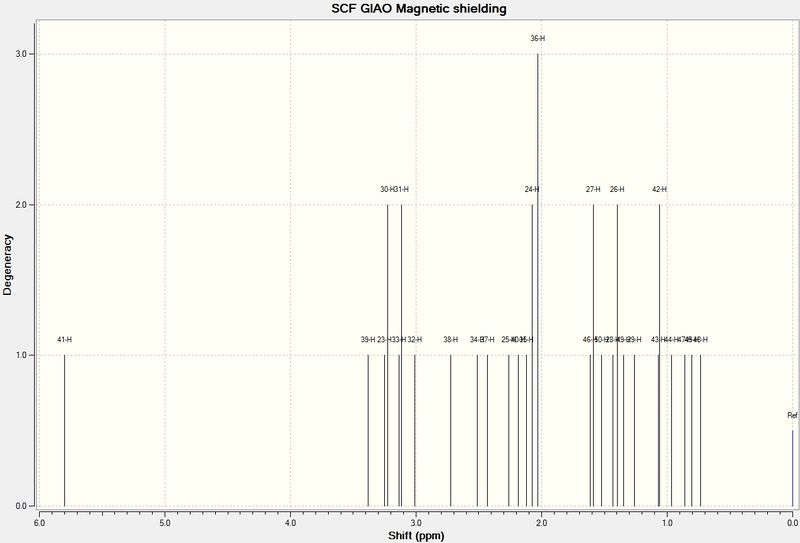
1H
These spectra actually have dramatically different styles. The literature spectrum is, as expected, less resolved. It also shows coupling, while the computed spectrum does not seem to represent the coupling of the system. Results still agreed relatively well. Shielding and deshielding is also affected by angle rather than simply the atoms nearby. This is shown in distinct values for members of the same methyl group.
| Hydrogen | Predicted Chemical Shift (ppm) | Literature Chemical Shift (ppm) |
|---|---|---|
| Alkene Proton | 5.80 | 4.84 |
| 1,3/1,4 Relationship with S | 3.38, 3.25, 3.23, 3.11 | 3.4-3.1 |
| H antiperiplanar to S | 3.01 | 2.99 |
| Hs around 6 member-ring (relative shielding affected by proximity and angle of S) | 2.72-1.4 | 2.8-1.34 |
| Hydrogen alpha to carbonyl | 2.5 | not resolved |
| Methyl group beta to carbonyl | 1.26 | 1.25 |
| Bridging methyl group between ketone and alkene | 1.07 and 1.06 | 1.1, 1.004 |
| Methyl Environments | 0.74-1 | 0.8-1 |
Literature Molecule - A Cyclopropane and its Isomer
A paper on the subject of Ruthenium catalysed cyclopropanations reports a mixture of isomers in the reaction shown below DOI:10.1021/jo034841a . The two products are the desired cyclopropane(46%) and a regioisomer - a cycloheptadiene(38%).
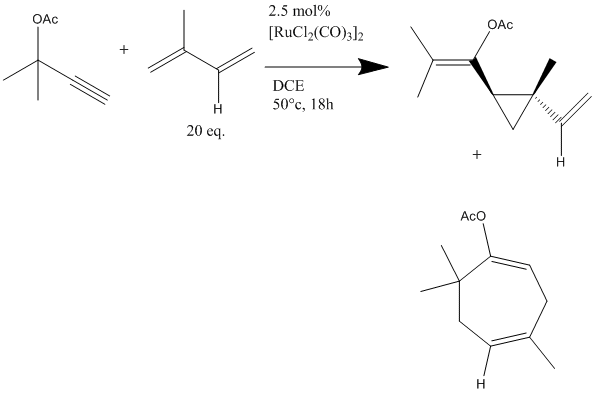
Despite being isomeric, these compounds are considerable different, and as such almost any technique could be used to differentiate between them. For example, NMR would be expected differ somewhat as a result of different connectivity in the molecules, similarly IR may differ, as despite the same functional groups, the molecules' shapes and symmetry will be different, affecting the molecules' vibrational modes.
Cyclopropane
The optimised MM2 structure shows a trisubstituted cyclopropane, with the 2 larger substituents sitting trans to each other. This molecule has 2 chiral centres, and the MM2 optimisation appears to give (S,S) stereochemistry.
Optimised Conformation of Product
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 2: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 1.1229 Bend: 4.4548 Stretch-Bend: 0.0480 Torsion: 5.0004 Non-1,4 VDW: -3.2740 1,4 VDW: 7.0612 Dipole/Dipole: 2.2605 Total Energy: 16.6738 kcal/mol Calculation completed ------------------------------------
This was then optimised in Gaussian, using a 6-31G (d,p)(B3LYP) basis set. DOI:10042/23076
| Paramater | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RmPW1PW91 |
| Basis Set | 6-31G(d,p) |
| Final Energy(au) | -618.39223446 |
| Gradient | 0.00000529 |
| Dipole Moment | 1.8574 |
| Point Group | C1 |
| Calculation Length | 2hr 14min 53s |
Cycloheptadiene
Unsurprisingly, as it is a 7-member ring system, both alkenes are cis, and the ester substituted carbon is an alkene, giving an enolate like structure. It should be noted that unlike the cyclopropane isomer, this molecule is achiral, and the main differences in conformation are likely to come from the orientation of the acetyl group or different ring conformations.
Optimised Conformation of Product
------------MM2 Minimization------------ Warning: Some parameters are guessed (Quality = 1). Iteration 54: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 1.5551 Bend: 4.8347 Stretch-Bend: 0.2166 Torsion: 5.1193 Non-1,4 VDW: -2.8812 1,4 VDW: 9.7360 Dipole/Dipole: 3.1974 Total Energy: 21.7778 kcal/mol Calculation completed ------------------------------------
This molecule was then analysed in Gaussian similar to above. Due to the limitation on cycles, the optimisation appears incomplete in log file. DOI:10042/23131
| Paramater | |
|---|---|
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RmPW1PW91 |
| Basis Set | 6-31G(d,p) |
| Final Energy(au) | -618.41759428 |
| Gradient | 0.00000529 |
| Dipole Moment | 4.2929 |
| Point Group | C1 |
| Calculation Length | 3hr 24min 12s |
Energy Comparison
This is an area of some confusion. From MM2 calculations, the cyclopropane isomer is more stable, in line with the experimental result. The gaussian optimisation, however, gave an energy difference of 15.9kcal/mol in favour of the cycloheptadiene, and the IR free energies below show a much smaller preference for the cycloheptadiene isomer. These results are puzzling, and suggest the reaction may be under kinetic control, or that there is an error in optimisation.
Cyclopropane NMR
13C
This spectrum, although at some points deviating by up to 5 ppm, displays a very similar trend, in the spacing of data and also in its chemical shift, to the reported literature molecule.
| Carbon | Predicted Chemical Shift (ppm) | Literature Chemical Shift (ppm) |
|---|---|---|
| Ester C | 164.4 | 169.2 |
| Monosubs. alkene | 139.5 | 145.2 |
| Tetrasubs. alkene - C bonded to O | 138.9 | 139.2 |
| Tetrasubs. alkene - C bonded to 2Me | 124.3 | 122.7 |
| Monosubs. alkene terminal C | 109.70 | 110.2 |
| Cyclopropane - bound to H and enolate | 32.2 | 27.0 |
| Cyclopropane - bound to Methyl & alkene | 31.0 | 25.3 |
| OAc methyl C | 21.3 | 20.5 |
| Cyclopropane bound Methyl | 21.1 | 19.5 |
| Methyl attached to alkene | 19.9 | 18.7 |
| Methyl attached to alkene | 18.5 | 17.6 |
| Cyclopropane Methylene group | 14.8 | 16.3 |
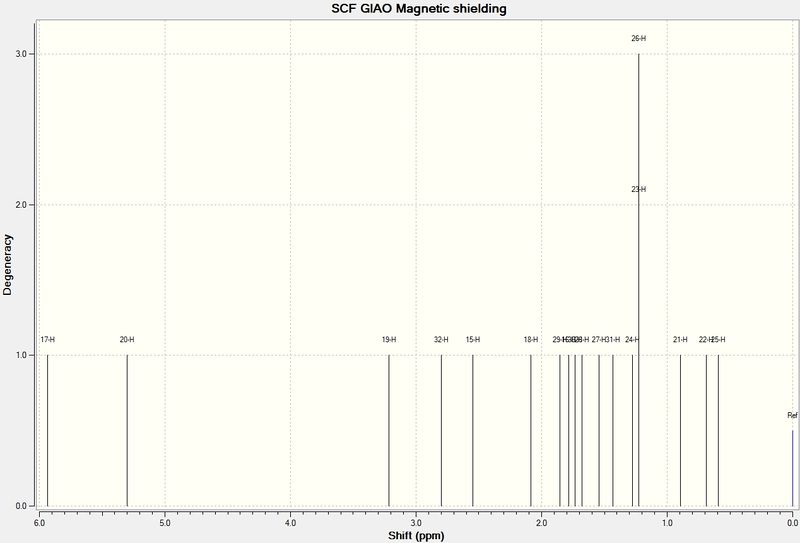
1H
Coupling Calculation DOI:10042/23295
The agreement between literature and calculated proton NMR spectra is in fact severely limited. Some agreement is noted, for example, the alkene protons, although differing by a relatively large value of ~1 ppm, show the same coupling and as such the same position in the spectrum. There are several sources of this disagreement. First of all, resolution of the computed spectrum is normally superior. This means that a larger distinction between chemical environments is possible. It would appear, however, that many protons grouped together are merely so because they have a similar chemical environment with regards to shielding, but in fact have different connectivity and may be quite disparate with regards to their position in the molecule or even subtle changes of dihedral angle. Furthermore, the NMR calculation proceedure is very sensitive to conformation, and unfortunately there is insufficient time to model even the few available conformations of the molecule.
| Hydrogen | Predicted Chemical Shift (ppm) | Literature Chemical Shift (ppm) |
|---|---|---|
| Alkene proton(beta to cyclopropane) | 6.72(1H, J=11,17 Hz) | 5.52 (1H, 11, 17 Hz) |
| Alkene protons (terminal) | 5.25 (2H, J=12,17 Hz) | 4.97 (1H, J=11 Hz), 4.92, J=17 Hz |
| Acetyl Methyl group | 2.29 ,2.25, 2.15 (4H) | 2.13 (3H) |
| Cyclopropane C-H | 1.71 (1H) | 1.78-1.89 (1H) |
| C-H(anitperiplanar to O) | 1.66 (4H) | 1.70 (3H) |
| Methyl group bound to alkene | 1.42 (2H) | 1.61 (3H) |
| Cyclopropane H | 1.32 (1H, J=10Hz, 2J=7Hz) | 1.09 (3H) |
| Cyclopropane bound methyl | 1.25 (2H, 2J=15 Hz) | 0.94 (1H, J= 4.8, 6.3 Hz) |
| Cyclopropane methylene or cyclopropane bound methyl | 0.43 (2H, J=0 Hz) | 0.66 1H, J=6.3, 8.1 Hz) |
Cycloheptadiene NMR
13C
As above, the carbon-13 NMR corresponds very well to literature. Although the chemical shifts are largely similar, which is to be expected because of the similar funtionality and therefore similar shielding of carbon atoms.
Arguably, however, there is sufficient difference to distinguish the 2 isomers simply by looking at those resonances where the structures diverge. Most notably, the methylene group on the cycloheptadiene ring can be compared to that of the cyclopropane, which is less deshielded, the cycloheptadiene and cyclopropane methylenes have shifts of 39.5 and 14.8 respectively. The peak at 39.5 could be particularly key as there are no peaks in the cyclopropane isomer's spectrum in either literature or prediction.
| Carbon | Predicted Chemical Shift (ppm) | Literature Chemical Shift (ppm) |
|---|---|---|
| Carbonyl | 164.8 | 169.7 |
| C-OAc | 156.9 | 154.4 |
| Alkene C | 141.4 | 140.4 |
| "" | 122.6 | 122.3 |
| "" | 113.9 | 114.5 |
| Quaternary C | 40.8 | 38.9 |
| Methylene group on ring | 39.5 | 38.6 |
| "" | 30.8 | 29.4 |
| Me | 28.7 | 27.0 |
| Me | 26.6 | 26.8 |
| Alkene bound Me | 25.9 | 24.7 |
| OAc Methyl C | 18.3 | 21.1 |
1H
Coupling Calculation - DOI:10042/23296
The problems already elaborated for the cyclopropane isomer and its 1H NMR are still applicable here. As mentioned above, the conformation of the ring and orientation of acetyl group are open to conformational differences. Similarly to above, however, there are resonances where the spectra certainly seem to agree, but also peaks in the computed spectrum absent from the literature. This highlights the superiority of the technique of modelling and its sincreased sensitivity. Again, a key point in the spectrum useful for distinction is δ=3.15 ppm. This represents the protons between the alkene groups which are β to each other. As a consequence of greater seperation of the alkenes in the cyclopropyl isomer, such a resonance is not observed and in fact, as in the carbon-13 example above, there is considerable space around this chemical shift value, allowing it to be analysed rather easily.
| Hydrogen | Predicted Chemical Shift (ppm) | Literature Chemical Shift (ppm) |
|---|---|---|
| Alkene proton | 5.84(1H, J=6.9 Hz) | 5.54 (1H, J=7.2 Hz) |
| Alkene proton(enolate) | 5.26(1H, J=14 Hz) | 5.22 (1H, J=6.4 Hz) |
| C-H between alkenes | 3.15 (1H,J=0 ) | - |
| C-H of acetyl methyl group | 2.78 (1H, J=0Hz) | 2.72 (2H, J=6.4Hz) |
| Alpha to alkene | 2.42 (1H, J=6.9 Hz) | 2.15 (2H, J=7.2 Hz) |
| Other H on methylene group between alkenes | 2.07(1H) | 2.12 (3H) |
| Methylene | 1.79 (1H) | 1.77 (3H) |
| Deshielded Me | 1.64 (4H) | - |
| Methyl groups | <1.4 (6H) | 1.02 (6H) |
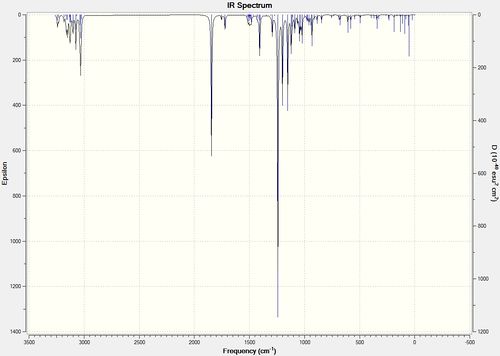
IR
IR spectra were predicted for both isomers by carrying out an optimisation and frequency calculation. This revealed an energy difference in the isomers of 10 kcal/mol(~42 kJ/mol). Perhaps unsurprisingly, the most IR active modes in both spectra are the stretches involving the acetyl group. This functional group is likely to have the largest effect on the dipole moment of the molecule and as such the bond stretching is also likely to have the greatest effect on the overall dipole moment and hence the spectra. Both spectra have the same number of vibrational modes, but it is interesting to note that the cycloheptadiene isomer has more vibrational modes that are highly IR active. This may be because the ring structure gives some vibrations a larger cumulative effect on the dipole moment. It is interesting to note that the carbon oxygen single bond displays some double bond character in its stretches, and this sort of resonance may give stabilisation to both molecules. These vibrational modes are pictured below.
The spectra are characterised with the corresponding literature peaks. This is something of an arbitrary assignment, however, because literature is often not rigourous in IR assignments and only the carbonyl peaks are analysed. These spectra are still useful, however, as a 'fingerprint' of the molecules, confirming their presence. However, each molecule had 90 vibrational modes, and only the peaks that produced high intensity are characterised, making it more similar to literature, although it appears that different peaks appeared at different intensities in literature and prediction.
Cyclopropane
Sum of electronic and thermal Free Energies= -618.311466
| Vibration | Frequency (cm-1) | Intensity | Literature |
|---|---|---|---|
| C-H rocking | 1122 | 41 | 1121 |
| C-H wagging | 1151 | 105 | 1180 |
| C-C stretch(cyclopropane beta to acetyl) | 1200 | 104 | 1215 |
| C-O stretch | 1241 | 356 | 1369 |
| Acetyl methyl scissoring | 1407 | 48 | 1445 |
| C=O stretch | 1844 | 188 | 1754 |
| C-H(alkene bound methyl gr.) symmetric stretch | 3031 | 67 | 2917 |
| C-H(alkene bound methyl gr.) asymmetric stretch | 3074 | 42 |
Cycloheptadiene
Sum of electronic and thermal Free Energies= -618.327409
| Vibration | Frequency (cm-1) | Intensity | Frequency (cm-1) Literature |
|---|---|---|---|
| C-H rocking | 1019 | 39 | 1063 |
| C-H scissoring | 1099 | 49 | 1093 |
| C-H wagging | 1131 | 137 | |
| C-O stretch(ring bound O) | 1232 | 394 | 1210 |
| C-C(in ring) stretching | 1255 | 50 | |
| Acetyl methyl C-H stretching | 1410 | 54 | 1452 |
| C=O stretching | 1853 | 382 | 1759 |
| Methyl C-H symmetric stretch | 3022 | 34 | 2928 |
| Methyl C-H asymmetric stretch | 3110 | 34 | 1966 |
| Alkene C-H stretching | 3162 | 31 |
Optical Rotation
Optical rotation measurements for the cycloheptadiene isomer are not particularly useful. Firstly, the sign of the rotation is not considered usable or accurate unless its magnitude is over 100, clearly not the case here. Secondly, the molecule is in fact achiral, and therefore has a magnitude close to zero. The cyclopropane isomer produces a value of +152.84 degrees. This is over the threshold to make it a usable value. This leaves is with the conclusion that the cyclopropane isomer is the (+) isomer. Analysis of these results is somewhat artificial because they are not reported in the literature, which suggests that their product is a racemate. Therefore, these calculation have, in a way, taken their results a step further. In actuality, however, these results are not necessarily reliable. It is impossible to know if the computation has produced the correct result in the absence of experimental data, such as a crystal structure.
Cyclopropane - DOI:10042/23289
Cycloheptadiene - DOI:10042/23290
| Isomer | [Alpha](5890.0 A) |
|---|---|
| Cyclopropane | +152.84 deg. |
| Cycloheptadiene | -4.27 deg. |
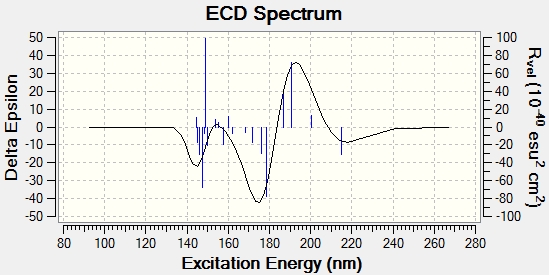
Circular Dischroism/UV-vis
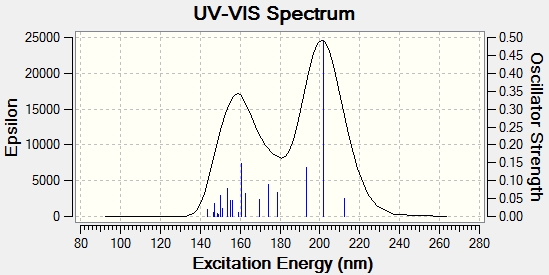
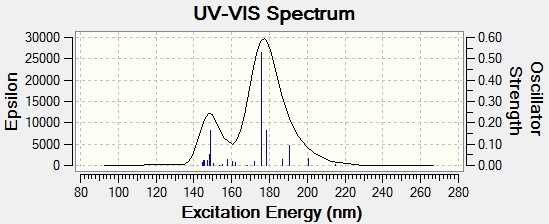
These spectra are included to give an analytical impression of the molecule. As they are absent from the literature, they are not very useful for comparitive purposes. It should be noted however, that the ECD spectrum of the cycloheptadiene isomer appears to show chirality, this may be when it is analysed in a specific conformation, when in fact it can interconvert by rotation, giving an average of these conformations, in other words they are present in an energy weighted average over time. Similarly, the UV spectra, which of course are analogous to UV spectra run without polarised light, are included to give an impression of the molecules and how their spectroscopic properties differ. The UV spectra show a higher λmax for cyclopropane, suggesting that it has a smaller band gap than the cycloheptadiene isomer.
Cyclopropane - DOI:10042/23292
Cycloheptadiene - DOI:10042/23291
CD
UV-vis
Conclusion
There are many inconsistencies in the data which suggest further analysis would be advisable, but the weight of evidence seems to confirm that an isomeric mixture of a substituted cyclopropane and a substituted cycloheptadiene. The percentages present are 48 and 36 respectively. A long time and a temperature of 50 degrees suggests thermodynamic control, but kinetic control is more likely based on the calculations, which is something that could be further investigated in the lab by altering the temperature or computationally by computing with a better basis set. There is emphasis at some points of how each isomer could be analysed by highlighting differences in the spectra. For example, in the Carbon-13 analysis. This means that it can be easily shown whether or not the product possesses the cycloheptadiene side product as an impurity. Another interesting calculation would have been Mass Spectrometry. While the literature only quotes the M+, it would be interesting to observe how the fragmentation patterns differ and how this can be used to distinguish between the isomers.