Rep:Mod:PJX4349
NH3 Analysis
summary information
- What is the molecule?
NH3.
- What is the calculation method?
RB3LYP.
- What is the basis set?
6-31G(d,p)
- What is the final energy E(RB3LYP) in atomic units (au)?
-56.55776873 au.
- What is the RMS gradient?
0.00000485 au.
- What is the point group of your molecule?
C3V.
geometric information
- the optimised N-H bond distance
1.01798 Å.
- the optimised H-N-H bond angle
105.741 degrees.
*bond angle comparison
The bond angle calculated from the optimized model is 105.741 degrees but the literature value found is 106.7 degrees.[1] Because the optimized model is calculated from the first principle assuming that the molecule we are looking at is the only one in the universe. But in reality there will always be some ammonia molecules around the one that we are looking at. The interactions between the molecules will affect the measuring of the actual bond angle and therefore there will be differences between the model value and the experimentally measured value.
"Item" table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986283D-10
Optimization completed.
-- Stationary point found.
finished .log file upload
test molecule |
The optimisation file is liked to here
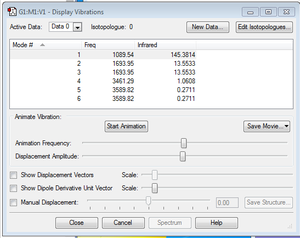
Animating the vibrations
- how many modes do you expect from the 3N-6 rule?
N=4. 3N-6=6 modes.
- which modes are degenerate (ie have the same energy)?
Mode 2 and 3, 5 and 6.
- which modes are "bending" vibrations and which are "bond stretch" vibrations?
Modes 1,2 and 3 are "bending" vibrations and modes 4,5 and 6 are "bond stretch" vibrations.
- which mode is highly symmetric?
Mode 4.
- one mode is known as the "umbrella" mode, which one is this?
Mode 1.
- how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2. Although there are 4 different peaks, two of them (with value 1.0608 and 0.2711) are too small to be seen.
Charge Analysis
- Write a sentence saying what charge (positive or negative) you would expect for N and H and why.
N should be negative and Hs should be positive. N will pull electron density towards itself because N is more electronegative than H.

N2 Analysis
summary information
- What is the molecule?
N2.
- What is the calculation method?
RB3LYP.
- What is the basis set?
6-31G(d,p)
- What is the final energy E(RB3LYP) in atomic units (au)?
-109.52412868 au.
- What is the RMS gradient?
0.00000365 au.
- What is the point group of your molecule?
D∞H.
geometric information
- the optimised N≡N bond distance
1.10550 Å.
"Item" table
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.000002 0.001800 YES
RMS Displacement 0.000003 0.001200 YES
Predicted change in Energy=-1.248809D-11
Optimization completed.
-- Stationary point found.
finished .log file upload
test molecule |
The optimisation file is liked to here
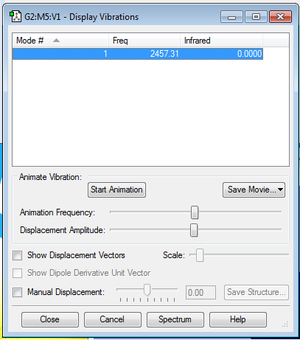
Animating the vibrations
The table below shows that there are no negative frequencies.
H2 Analysis
summary information
- What is the molecule?
H2.
- What is the calculation method?
RB3LYP.
- What is the basis set?
6-31G(d,p)
- What is the final energy E(RB3LYP) in atomic units (au)?
-1.17853930 au.
- What is the RMS gradient?
0.00012170 au.
- What is the point group of your molecule?
D∞H.
geometric information
- the optimised H-H bond distance
0.74309 Å.
"Item" table
Item Value Threshold Converged?
Maximum Force 0.000211 0.000450 YES
RMS Force 0.000211 0.000300 YES
Maximum Displacement 0.000278 0.001800 YES
RMS Displacement 0.000393 0.001200 YES
Predicted change in Energy=-5.852867D-08
Optimization completed.
-- Stationary point found.
finished .log file upload
test molecule |
The optimisation file is liked to here
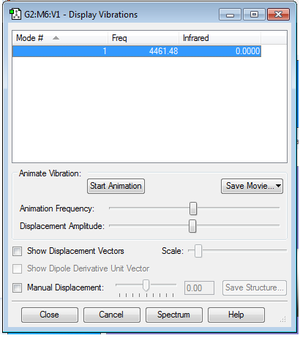
Animating the vibrations
The table below shows that there are no negative frequencies.
Energy Question
E(NH3)=-56.55776873 au
2*E(NH3)=-113.11553746 au
E(N2)=-109.52412868 au
E(H2)=-1.17853930 au
3*E(H2)=-3.5356179 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.05579088 au
the energy for converting hydrogen and nitrogen gas into ammonia gas:
ΔE=-0.05579088 au=-146.478966598 KJ/mol=-146.48 KJ/mol (2d.p.)
The ammonia product is more stable because the formation of ammonia is an exothermic reaction and therefore the product is lower in energy than the reactants.
CH4 Analysis
summary information
- What is the molecule?
CH4.
- What is the calculation method?
RB3LYP.
- What is the basis set?
6-31G(d,p)
- What is the final energy E(RB3LYP) in atomic units (au)?
-40.52401405 au.
- What is the RMS gradient?
0.00002267 au.
- What is the point group of your molecule?
TD.
geometric information
- the optimised C-H bond distance
1.09194 Å.
- the optimised H-C-H bond angle
109.471 degrees.
"Item" table
Item Value Threshold Converged?
Maximum Force 0.000044 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000124 0.001800 YES
RMS Displacement 0.000066 0.001200 YES
Predicted change in Energy=-1.089024D-08
Optimization completed.
-- Stationary point found.
finished .log file upload
test molecule |
The optimisation file is liked to here
Vibrations Analysis
Following the 3N-6 rule (in this case N=5), we expect 3N-6=9 modes. Mode 1,2,3, mode 4,5, and mode 7,8,9 are degenerate. Modes 1 to 5 are bending vibrations and modes 6 to 9 are bond stretch vibrations. There is no changes in dipole moment in modes 4,5 and 6 therefore they are infrared inactive and this results in 0 reading in the infrared spectrum. There are two peaks in the infrared spectrum.


Charge Analysis
C should be negative and Hs should be positive. C will pull electron density towards itself because C is more electronegative than H. But the molecule itself is neutral.

Molecular Orbital Analysis
The picture MO1. below shows the 1s orbital of C atom. The 1s orbital of C atom is not bonding with any other atomic orbitals so it makes no contribution to the MO.
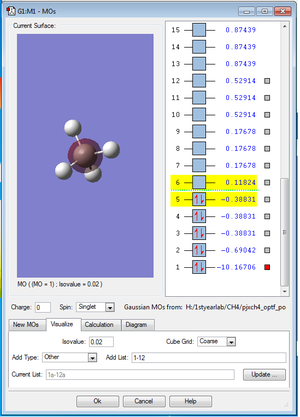
The picture MO2. below shows the bonding σs molecular orbital formed by the 2s atomic orbital of C and 1s atomic orbital of H. It is occupied by 2 electrons and contributes two bonding electrons to the bond order.

The picture MO3. below shows the bonding σx molecular orbital formed by the 2px atomic orbital of C and 1s atomic orbital of H. There are two other MOs with the same energy but formed by the 2py and 2pz atomic orbital of C. These three degenerate orbitals are the HOMO. They are all occupied by two electrons and therefore contribute 6 bonding electrons to the bond order. From the Valence bond theory we know that sp3 hybridization of 2s and 2p AOs of C produces 4 degenerate MOs but we do not see any evidence here using the optimization model.

The picture MO4. below shows the anti-bonding σs* molecular orbital formed by the 2s atomic orbital of C and 1s atomic orbital of H. The σs* MO is the LUMO. It is empty and therefore contributes no anti-bonding electrons to the bond order.
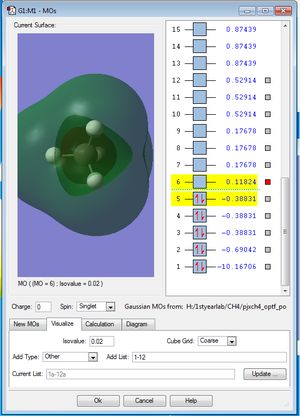
The picture MO5. below shows the anti-bonding σx* molecular orbital formed by the 2px atomic orbital of C and 1s atomic orbital of H. There are two other anti-bonding MOs with the same energy but formed by the 2py and 2pz atomic orbital of C. They are all quite high in energy. These three anti-bonding MOs are all empty and therefore contribute no anti-bonding electrons to the bond order.

There are 8 bonding electrons in total and 0 anti-bonding electrons so the bond order is 4.
Reference
[1] the bond angle of ammonia: CRC Handbook of Chemistry and Physics, 96th ed. http://www.hbcpnetbase.com/ Section 9, page 9-26
