Rep:Mod:Mvemjsun8
NH3 Optimisation
General Information
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy or E(RB3LYP) | -56.55776873 a.u. |
| RMS Gradient Norm | 0.00000485 a.u. |
| Dipole Moment | 1.8466 Debye |
| Point Group | C3V |
| Average N-H Bond Length | 1.02 Å |
| Average H-N-H Bond Angle | 106° |
Item Table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
NH3 Optimised |
Link to NH3 Log File: https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:Tahmidur_NH3_OPTIMISATION_Data_1.LOG
Vibrations
| Mode Number | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Wavenumber / cm-1 | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| Symmetry | A1 | E | E | A1 | E | E |
| Intensity / a.u. | 145 | 14 | 14 | 1 | 0.3 | 0.3 |
| Image | 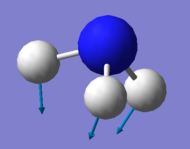
|
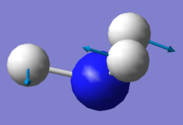
|
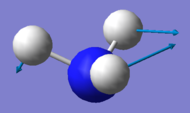 |
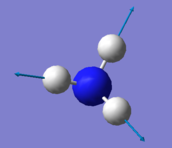
|

|
|
More Info
| Modes of Vibrations | 3N - 6 = 3(4) - 6 = 6 Modes of Vibration |
| Degenerate Modes | Vibrations with 1694 cm-1 (Modes 2 & 3)
and 3590 cm-1 (Modes 5 & 6) |
| Bending Vibrations | Modes 1, 2 & 3 |
| Stretching Vibrations | Modes 4, 5 & 6 |
| Highly Symmetric Mode | Mode 4 |
| "Umbrella" Mode | Mode 1 |
| Estimated Experimental
Bands Shown |
3 Bands |
|---|
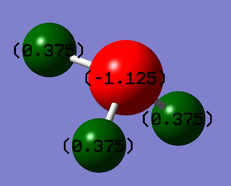
Charge Distrubution
It is expected that the charge of the nitrogen will be negative compared to the hydrogens, as they are more elcetronegative and thus have a larger electron density around them. The charge values of nitrogen are -1.125, and for hydrogen it is 0.375.
H2 Optimisation
General Information
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy or E(RB3LYP) | -1.17853936 a.u. |
| RMS Gradient Norm | 0.00000017 a.u. |
| Dipole Moment | 0 Debye |
| Point Group | D∞H |
| H-H Bond Length | 0.74 Å |
Item Table
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
H2 Optimised |
Link to H2 Log File: https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:Tahmidur_H2_OPTIMISATION.LOG
Vibrations
| Mode Number | 1 |
|---|---|
| Wavenumber / cm-1 | 4466 |
| Symmetry | SGG |
| Intensity / a.u. | 0 |
| Image | 
|
More Info
| Modes of Vibrations | 3N - 5 = 3(2) - 5 = 1 |
|---|---|
| Stretching Vibration | Mode 1 |
| Estimated Experimental
Bands Shown |
0 |
Charge Distribution
H2 is a simple diatomic molecule, and both atoms have the same electronegativity. Both have a charge of 0. Thus there is no overall dipole, and consequently a dipole can't be found in the vibration above.
N2 Optimisation
General Information
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy or E(RB3LYP) | -109.52412868 a.u. |
| RMS Gradient Norm | 0.00000060 a.u. |
| Dipole Moment | 0 Debye |
| Point Group | D∞H |
| N-N Bond Length | 1.11 Å |
Item Table
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
N2 Optimised |
Link to N2 Log File: https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:Tahmidur_N2_OPTIMISATION.LOG

Vibrations
| Mode Number | 1 |
|---|---|
| Wavenumber / cm-1 | 2457 |
| Symmetry | SGG |
| Intensity / a.u. | 0 |
| Image | 
|
More Info
| Modes of Vibrations | 3N - 5 = 3(2) - 5 = 1 |
|---|---|
| Stretching Vibration | Mode 1 |
| Estimated Experimental
Bands Shown |
0 |
Charge Distribution
N2 is a simple diatomic molecule, and both atoms have the same electronegativity. Both have a charge of 0. Thus there is no overall dipole, and consequently a dipole can't be found in the vibration above.
Mono-metallic TM complex with N2
The TM Complex found was trans-bis(1-(diethylphosphino)-N-((diethylphosphino)methyl)-N-(2,6-difluorobenzyl)methanamine)-bis(dinitrogen)-chromium, which has a Refcode of AQEZIH. The N2 molecule coordinates with a central chromium atom, along with another N2 in the axial positions, and the 4 equatorial groups give it an octahedral structure. This bond length for N2 is 1.127 Å, whereas the calculated length is 1.106 Å to 4 sf. This could be due to the coordinating nitrogen having a postive charge, thus there is increase nuclear repulsion between the two nitrogen atoms.
Link to journal: http://dx.doi.org/10.1039/C6CC03449G
Link to strucure on CCDC: https://www.ccdc.cam.ac.uk/structures/search?sid=ConQuest&coden=CHCOFS&year=2016&spage=9343&volume=52&id=doi:10.1039/C6CC03449G&pid=ccdc:1445489
Relating to the Haber-Bosch Process
Determining energy of reaction
| Energy Calculation | Energy / a.u. | Energy / kJ.mol-1 |
|---|---|---|
| E(NH3)= | -56.55776873 | -148492.4 |
| 2 x E(NH3) = | -113.11553746 | -296984.8 |
| E(N2)= | -109.52412868 | -287555.6 |
| E(H2)= | -1.17853936 | -3094.3 |
| 3 x E(H2)= | -3.53561808 | -9282.8 |
| ΔE= 2 x E(NH3) - [ E(N2) + 3 x E(H2) ] = | -0.05579076 | -146.5 |
The gaseous product (ammonia) is more stable as they are in lower in energy than the gaseous products due to the forward reaction being exothermic, as it releases energy during the reaction. Also, as there are more moles of gas in the products, the entropy would be higher in the products than the reactants, as there are more ways of arranging the molecules.
SbF5 Optimisation
General Information
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | DGDZVP |
| Final Energy or E(RB3LYP) | -6814.68819375 a.u. |
| RMS Gradient Norm | 0.00004712 a.u. |
| Dipole Moment | 0 Debye |
| Point Group | D3H |
| Average Sb-F Bond Length | (1.91328 x 3 + 1.92044 x 2) / 5 = 1.92Å |
| F-Sb-F Bond Angles | 90° and 120° |
Item Table
Item Value Threshold Converged? Maximum Force 0.000118 0.000450 YES RMS Force 0.000042 0.000300 YES Maximum Displacement 0.000466 0.001800 YES RMS Displacement 0.000164 0.001200 YES
SbF5 Optimised |
Link to SbF5 Log File: https://wiki.ch.ic.ac.uk/wiki/index.php?title=File:Tahmidur_SBF5_OPTIMISED_DGDZVP.LOG
Vibrations
Modes 1-6
| Mode Number | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Wavenumber / cm-1 | 89 | 89 | 229 | 229 | 238 | 238 |
| Symmetry | E' | E' | E' | E' | E" | E" |
| Intensity / a.u. | 0 | 0 | 52 | 52 | 0 | 0 |
| Image | 
|

|
 |
|
|
|
Modes 7-12
| Mode Number | 7 | 8 | 9 | 10 | 11 | 12 |
|---|---|---|---|---|---|---|
| Wavenumber / cm-1 | 254 | 594 | 612 | 669 | 669 | 671 |
| Symmetry | A2" | A1' | A1' | E' | E' | A2" |
| Intensity / a.u. | 52 | 0 | 0 | 83 | 83 | 107 |
| Image | 
|

|
 |
|
|

|
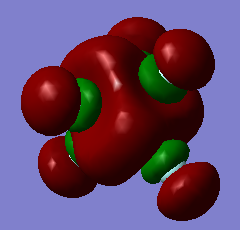
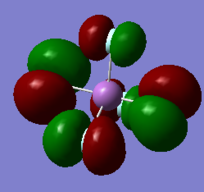
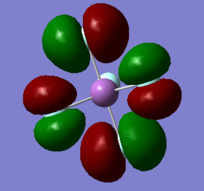
More Info
| 'Modes of' Vibrations | 3N - 6 = 3(6) - 6 = 12 Modes of Vibration |
| Degenerate Modes | Vibrations with 89 cm-1 (Modes 1 & 2), 229 cm-1 (Modes 3 & 4),
238 cm-1 (Modes 5 & 6) and 669 cm-1 (Modes 10 & 11) |
| Bending Vibrations | Modes 1-7 |
| Stretching Vibrations | Modes 8-12 |
| Symmetric Modes | Mode 8 & 9 |
| Experimental Bands Shown | 4 Bands |
|---|
Charge Distrubution
It is expected that the charge of the fluorines will be negative compared to the antimony, as they are more elcetronegative and thus have a larger electron density around them. The charge values of nitrogen are -0.626 for 3 atoms and -0.637 for 2 atoms. Antimony itself has a charge of 3.152. The molecule is too symmetrical for any permenant dipole to form, though the bonding is polar.
Molecular Orbitals Examined
MO 48 HOMO
The molecular orbital 48 represents the HOMO of SbCl5, which is formed of 3p atomic orbitals that are not in phase and don't symmetrically aline, thus representing the non-bonding of the fluorines. Being the HOMO, it is the last occupied orbital. The electrons in this orbital would allow it to act as a nucleophile, and have an energy of -0.45219 a.u.
MO 49 LUMO
In comparison, the molecular orbital 49 is the LUMO, formed of 5d orbitals of SB and the 2p orbitals of fluorine. These are antibonding and consequently out-of-phase. This is much higher in energy than the HOMO ( -0.45219 a.u.).
MO 47 HOMO-1
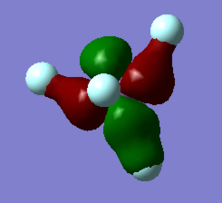
This molecular orbital is next one lower in energy than the HOMO, and is simply the 2p orbitals of fluorine that can't be symmetrically combined, and are therefore non-bonding. They are perpendicular to the ones found in the HOMO, and actually have the same energy (-0.45219 a.u.). It stands to reason that these are actually the same energy, and neither one is preferrred as the HOMO because of this. They can be matched to either 2pz or 2py respectively.
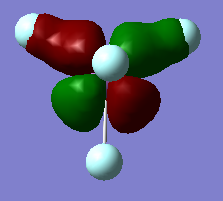
MO 25 HOMO-23 and MO 26 HOMO-22
This 25th molecular orbital is much deeper in energy than the HOMO (sitting at -1.61965 a.u.). It is a combination of s atomic orbitals that symmetrically line up with the d orbital of the fluroine. This is an in-phase bonding orbital. The same bonding occurs in MO 26 HOMO-22, where there is a orthogonal d orbital that combines/mixes with the s atomic orbitals of fluorine.
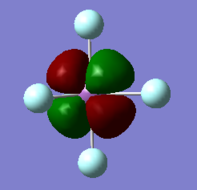
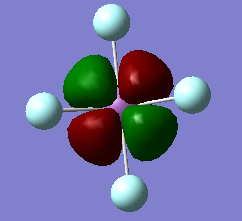
MO 27 HOMO-21 and MO 28 HOMO-20
The 27th molecular orbital is solely a d orbital that is non-bonding (as it's symmetry doesn't match with any other atomic orbitals). It has an energy of -1.61097 a.u., and is degenerate with MO 28 HOMO-20, which is the perpendicular d orbital and is also non-bonding.
Independant Work
Missing H-H Bond
When optimising the H2 molecule using calculation method of RB3LYP and base set of 6-31g(d,p), the hydrogen doesn't appear to have any bond in between the atoms. Although this may seem strange, guassian has still succefully optimised your molecule (see items table). It could be that the H-H bond length is longer than the average or maximum guassians parameter are set at to draw the bond in. The data is still valid, and most other data such as bond length is accurately predicted to a degree. This may occur for other atoms bonded to H where the mathematics for guassian decide not to draw the bond in the geometry even though there definitely should be a bond there according to current scientific knowledge.
Link to reasoning: https://www.researchgate.net/post/Why_bonds_breaks_after_the_optimization_in_Gaussian09
Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES, however you claimed to expect 3 experimental bands and give no reasons for this number. You correctly stated that there are two sets of degenerate modes - this explains a spectrum with 4 peaks. However there are only 2 peaks visible as peaks 4, 5 and 6 are of too low an intensity to be visible.
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES, you could have explained that the charges are 0 as the electronegativities are equal.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES - however you state a positive charge on the nitrogen to be the reason for an increase in NN-bond length. You should have discussed a decrease of the electron density between the N atoms which leads to a lower bond strength and higher bond length.
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
You have done a good job of presenting this information, very well done! You calculated Sbf5 and in the section discussing the charge distribution you stated : 'The charge values of nitrogen are -0.626 for 3 atoms and -0.637 for 2 atoms'.This sentence does not make sense at all as the molecule is not containing nitrogen!
Independence 0.5/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far
You have included a discussion of a technical issue occurring in gaussian - great! However you basically summarised a discussion in an online forum. For a full mark this section should be based on your own results.
