Rep:Mod:MinqiPan5318
NH3
logfile of ammonia optimisation
Optimisation Summary
| ammonia | |
|---|---|
| Molecular Formula | NH3 |
| Calculation Method | RB3LYP |
| Basic Set | 6-31G(d,p) |
| Final Energy | -56.55776873 a.u. |
| RMS Gradient | 0.00000485 a.u. |
| Point Group | C3V |
| bond length | 1.01798 Å |
| bond angle | 105.7° |
Optimisation Parameters
'''Item Value Threshold Converged?''' Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986270D-10 Optimization completed. -- Stationary point found. ---------------------------- ! Optimized Parameters ! ! (Angstroms and Degrees) ! -------------------------- -------------------------- ! Name Definition Value Derivative Info. ! -------------------------------------------------------------------------------- ! R1 R(1,2) 1.018 -DE/DX = 0.0 ! ! R2 R(1,3) 1.018 -DE/DX = 0.0 ! ! R3 R(1,4) 1.018 -DE/DX = 0.0 ! ! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 ! ! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 ! ! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 ! ! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 ! --------------------------------------------------------------------------------
ammonia molecule |
Modes of Vibrations
| wavenumber cm-1 | 1089.54 | 1693.95 | 1693.95 | 3461.29 | 3589.82 | 3589.82 |
|---|---|---|---|---|---|---|
| symmetry | A1 | E | E | A1 | E | E |
| intensity (arbitary units) | 145.3814 | 13.5533 | 13.5533 | 1.0608 | 0.2711 | 0.2711 |
| image |  |
 | | |
 |
 |
 |
|
Answers of the questions
how many modes do you expect from the 3N-6 rule?
6
which modes are degenerate (ie have the same energy)?
modes of frequecy 1693.95 are degenerated; modes of frequecy 3589.82 are degenerated.
which modes are "bending" vibrations and which are "bond stretch" vibrations?
modes of frequecy 1693.95 and 1089.54 are bending; modes of frequecy 3589.82 and 3461.29 are stretching
which mode is highly symmetric?
mode of frequecy 3461.29
one mode is known as the "umbrella" mode, which one is this?
mode of frequecy 1089.54
how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
4, at frequencies of 1089.54,1693.95 , 3461.29, 3589.82. However, the peaks at 3589.82 and 3491.29 will have much more lower freqencies than others.
Distribution of Charge
charge on N atom: -1.125
charge on H atom: 0.375
Nitrogen is more electronegative than Hydrogen, so it has a more negative charge, i.e. more electron density on it
H2
logfile of hydrogen optimisation
Optimisation Summary
| hydrogen | |
|---|---|
| Molecular Formula | H2 |
| Calculation Method | RB3LYP |
| Basic Set | 6-31G(d,p) |
| Final Energy | -1.17853936 a.u. |
| RMS Gradient | 0.00000017 a.u. |
| Point Group | D*H |
| Bond length | 0.74309 Å |
| Bond angle | 180° |
Optimisation Parameters
Item Value Threshold Converged?
Maximum Force 0.000211 0.000450 YES
RMS Force 0.000211 0.000300 YES
Maximum Displacement 0.000278 0.001800 YES
RMS Displacement 0.000393 0.001200 YES
Predicted change in Energy=-5.852867D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7431 -DE/DX = -0.0002 !
--------------------------------------------------------------------------------
hydrogen molecule |
Modes of Vibrations
| wavenumber cm-1 | 4461.48 |
|---|---|
| symmetry | SGG |
| intensity (arbitary units) | 0 |
| image | 
|
Distribution of Charge
charge on H atom: 0
Becuase the molecule is made of two identical atoms with the same electronegativity, the molecule is nuetral
N2
logfile of nitrogen optimisation
Optimisation Summary
| nitrogen | |
|---|---|
| Molecular Formula | N2 |
| Calculation Method | RB3LYP |
| Basic Set | 6-31G(d,p) |
| Final Energy | -109.52412868 a.u. |
| RMS Gradient | 0.00000060 a.u. |
| Point Group | D*H |
| Bond length | 1.10550 Å |
| Bond angle | 180° |
Optimisation Parameters
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.000002 0.001800 YES
RMS Displacement 0.000003 0.001200 YES
Predicted change in Energy=-1.248809D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
nitrogen molecule |
Modes of Vibrations
| wavenumber cm-1 | 2457.31 |
|---|---|
| symmetry | SGG |
| intensity (arbitary units) | 0 |
| image | 
|
Distribution of Charge
charge on H atom: 0
becuase it is made by two identical atoms with the same electronegativity, and it's a neutual molecule.
C40H64CoN2P3Si
| Molecular Formula | C40H64CoN2P3Si |
| Unique identifier | VEJSEL |
| N-N bond distance I obtaines | 1.105(5) Å |
| N-N bond distance in the structure | 1.154(9)Å [1] |
The bond length of N-N in the metal complex is larger than that of N2 molecule. This is due to the interaction with other part of the metal weakens the bond between the two nitrogen atoms. Electron density between nitrogen atoms are spreaded out towards the metal. Also, Gaussian does not consider the actual bonding between the atoms, it only takes into account the relative position of the two atoms to obtain a minimun energy. Any other interactive would alter the calculation performed by Gaussian.
Harber-Bosch Process
E(NH3)=-56.55776873 a.u.
2*E(NH3)= -113.1155375 a.u.
E(N2)= -109.52412868 a.u.
E(H2)= -1.17853936 a.u.
3*E(H2)= -3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05577074 a.u. = -146.4 kJ/mol
CO2
logfile of carbon dioxide optimisation
Optimisation Summary
| hydrogen | |
|---|---|
| Molecular Formula | CO2 |
| Calculation Method | RB3LYP |
| Basic Set | 6-31G(d,p) |
| Final Energy | -188.58093945 a.u. |
| RMS Gradient | 0.06653680 a.u. |
| Point Group | D*H |
| Bond length | 1.25840 Å |
| Bond angle | 180° |
Optimisation Parameters
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000017 0.000300 YES
Maximum Displacement 0.000021 0.001800 YES
RMS Displacement 0.000015 0.001200 YES
Predicted change in Energy=-5.259645D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1691 -DE/DX = 0.0 !
! R2 R(1,3) 1.1691 -DE/DX = 0.0 !
! A1 L(2,1,3,-2,-1) 180.0 -DE/DX = 0.0 !
! A2 L(2,1,3,-3,-2) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
carbon dioxide molecule |
Modes of Vibrations
| wavenumber cm-1 | 639.99 | 639.99 | 1372.08 | 2436.37 |
|---|---|---|---|---|
| symmetry | SGG | PIU | PIU | SGU |
| intensity (arbitary units) | 30.7186 | 30.7186 | 0.0000 | 545.8344 |
| image |  |
 |
 |

|
Distribution of Charge
charge on C atom: 1.022
charge on O atom: -0.511
oxygen is more elecronegative so it has more electron density than carbon.
MO diagrams
MO=1
This is the 1st MO of CO2, with the lowest energy and degenerate with the 2rd MO. This orbital is made of two 1s orbitals on the oxygen, however, as the MOs are two small and two far apart, they barely overlap.
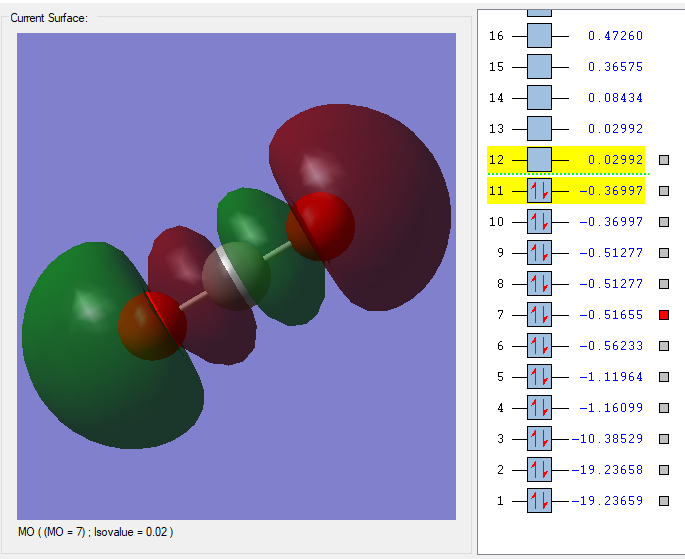
MO=7
This is the 7th MO of CO2. This orbital is mainly made by 2p orbitals on C and Os, but it also has some sp mixing. This is a mixture of bonding and antibonding orbitals, but with more antibonding-character.
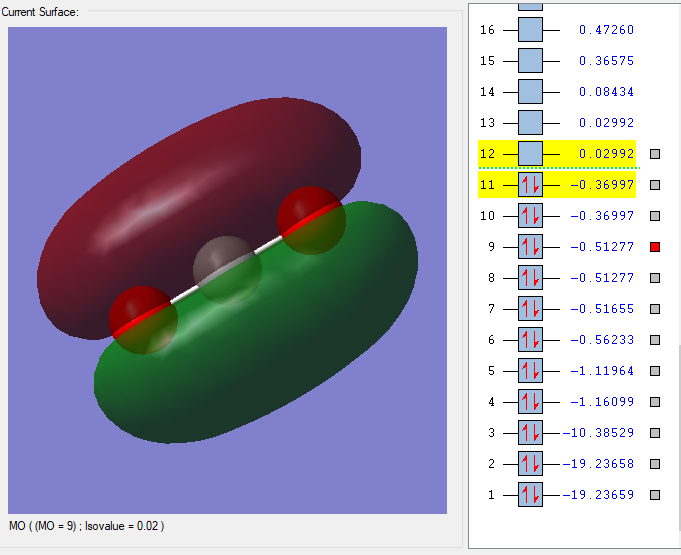
MO=9
This is the 9th MO of CO2, degenerate with the 8th MO. It is made by 2p orbitlas on C and Os. It is a bonding orbital, contribute to the pi bonding of the C-O bond. We can also see from the diagram that the pi electrons are delocalised along the whole molecule.
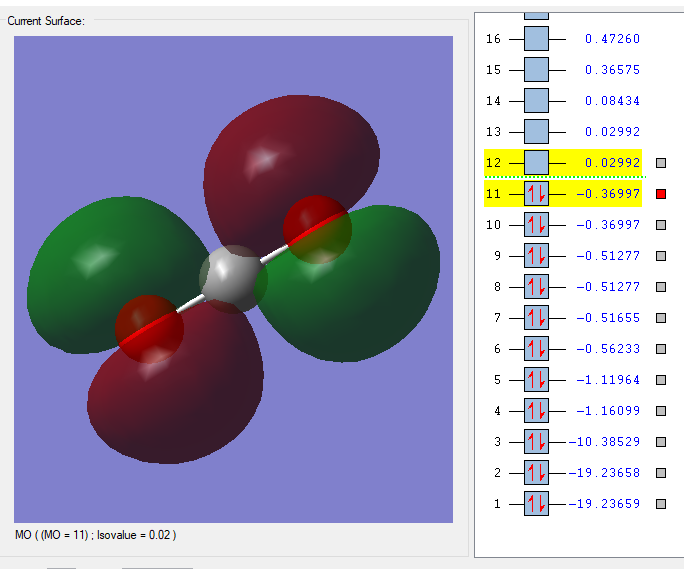
MO=11
This is the 11th MO of CO2, degenerate with the 10th MO. It is made by out of phase overlaping of 2p orbitals on the oxygem. It is antibonding.
MO=12
This is the 12th MO of CO2, also the LUMO of the molecule. This is made by out-of-phase overlapping of 2ps and also 3ps orbitals on C and Os. This is an antibonding orbitals but it is unfilled so does not contribute to the bonding of the molecule.
Independence
The energy of CO2 calculated by Gaussian is -188.58093945 a.u. = -495.1kJ/mol; the experimental data of enthalpy of formation of CO2 gas at standard conditions is -393.52kJ/mol[2] The literature value shows that the C=O bond is weaker than we would imagine, this is due to the interaction of CO2 molecules with other particles in the gas phase. It is impossible to measure the enthalphy of formation of a single molecule, thus some kind of molecular interaction will exist. However, Gaussian calculated molecular energy of a single isolated CO2 molecule. Intermolecular interaction tend to spread out electron density and therefore weaken the bonds. Therefore, the experimental value is smaller than the theoretical value.
References
- ↑ https://onlinelibrary.wiley.com/doi/full/10.1002/ejic.201700569.
- ↑ Chase, M.W., Jr., NIST-JANAF Themochemical Tables, Fourth Edition, J. Phys. Chem. Ref. Data, Monograph 9, 1998, 1-1951.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES, most answers are correct. However there are only 2 visible peaks in the spectra of NH3, due to the low intensity (not "frequency" as you stated) of the other 2 peaks. (See infrared column in vibrations table.)
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES, However you have given a bond angle of 180 for N2 and H2, there are no bond angles in diatomic molecules. Bond angles involve exactly 3 atoms.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES, good attempt at an explanation. This statement "Also, Gaussian does not consider the actual bonding between the atoms, it only takes into account the relative position of the two atoms to obtain a minimun energy." is wrong but it is beyond the scope of this lab to understand how Guassian and the method you applied work, you will learn this further on your course!
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, good explanations well done!
See this wiki page for more details on the CO2 MOs.
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
You checked a result against the literature well done!
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far