Rep:Mod:MMS4518
NH3 molecule
NH3 molecule |
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Point group: C3V
Final energy E(RB3LYP): -56.55776873 a.u.
RMS gradient: 0.00000485 a.u.
optimised N-H bond length: 1.02 A
optimised H-N-H bond angle: 106°
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
Vibrational Analysis
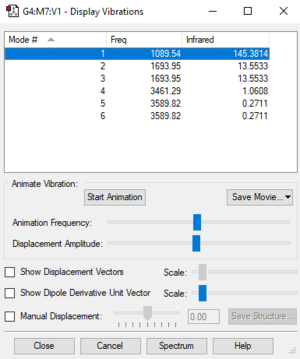
Number of modes expected from 3N-6 rule: 6
Degenerate modes: 2 and 3, 5 and 6
Bending vibrations: 1, 2, 3
Stretching vibrations: 4, 5, 6
Highly symmetric mode: 4
"Umbrella" mode: 1
Number of bands expected to see in an experimental spectrum of gaseous ammonia: 2 (Degeneracy means that 1 band is observed for 2 modes since they have the same frequency. Intensity of 4,5 and 6 are low)
| Wavenumber cm-1 | Symmetry | Intensity | |
|---|---|---|---|
| 1 | 1090 | A1 | 145 |
| 2 | 1694 | E | 13 |
| 3 | 1694 | E | 13 |
| 4 | 3461 | A1 | 1 |
| 5 | 3590 | E | 0 |
| 6 | 3590 | E | 0 |
IR intensities vary with the amount of change in dipole in the molecule. The first vibration has the strongest intensity because the dipole of the molecule changes the most in that vibration.
Charge Analysis
Charge on N atom: -1.125
Charge on H atoms: 0.375
Since nitrogen atoms are more electronegative than the hydrogen atom, a negative charge is expected on the nitrogen atom and positive charges are expected on the hydrogen atoms.
N2 molecule
N2 molecule |
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Point group: D∞h
Final energy E(RB3LYP): -109.52412868 a.u.
RMS gradient: 0.00000060 a.u.
Optimised bond length: 1.1 A
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
Vibrational Analysis
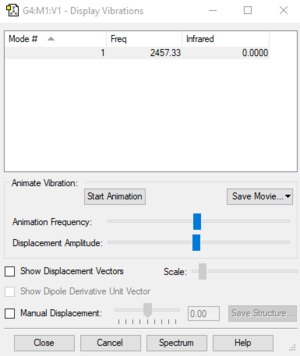
| Wavenumber cm-1 | Symmetry | Intensity |
|---|---|---|
| 2457 | SGG | 0 |
Number of Vibrations according to 3N-5: 1
Bending: None
Stretching: 1
IR Bands: 0 (as there is no change in dipole moment)
Charge Analysis
Charge on H atom: 0 (The molecule is nonpolar)
H2 molecule
H2 molecule |
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Point group: D∞h
Final energy E(RB3LYP): -1.17853936 a.u.
RMS gradient: 0.00000017 a.u.
Optimised bond length: 0.74 A
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
Vibrational Analysis
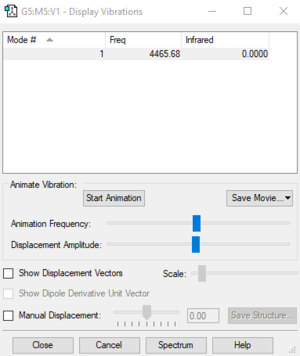
| Wavenumber cm-1 | Symmetry | Intensity |
|---|---|---|
| 4465 | SGG | 0 |
Number of Vibrations according to 3N-5: 1
Bending: None
Stretching: 1
IR Bands: 0 (as there is no change in dipole moment)
Charge Analysis
Charge on H atom: 0 (The molecule is nonpolar)
Transition Metal Complex
Identifier: NIHQIF [1]
N-N optimised bond distance: 1.10550 Å
N-N bond distance in complex: 1.131(8)Å [1]
The bond distances of the complex and the computational model are different as the one is experimentally measured and the other is an outcome from the optimisation calculation. When a ligand is bonded to a metal, sharing of electrons occurs. The interaction between the electrons in the N-N bond and the metal causes a lowering of electron density in the N-N bond, and therefore, the bond length increases due to the decreased electron to nucleus attraction. This also means that the N-N bond strength in the complex is weaker. A computational reason would be that the bond order in N2 and in the complex is different and there might be limitation to the optimisation.
Haber-Bosch process
The Haber-Bosch process [2] is an industrial process of which nitrogen gas and hydrogen gas are converted to ammonia. Ammonia is largely used in fertilisers today. The enthalpy calculation is as follow:
E(NH3)= -56.5577687 a.u.
2*E(NH3)= -113.1155374 a.u.
E(N2)= -109.5241286 a.u.
E(H2)= -1.1785393 a.u.
3*E(H2)= -3.5356179 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.0557909 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -146.5 kJ/mol
The exothermic reaction shows that ammonia is more stable than the reactants.
CO2 molecule
NH3 molecule |
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Point group: D∞h
Final energy E(RB3LYP): -188.58093945 a.u.
RMS gradient: 0.00001154 a.u.
Optimised C=O bond length: 1.17 A
Optimised O=C=O bond angle: 180°
Item Value Threshold Converged? Maximum Force 0.000024 0.000450 YES RMS Force 0.000017 0.000300 YES Maximum Displacement 0.000021 0.001800 YES RMS Displacement 0.000015 0.001200 YES
Vibrational Analysis
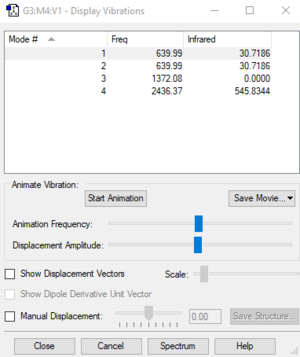
Number of modes expected from 3N-5 rule: 4
Degenerate modes: 1 and 2
Bending vibrations: 1 and 2
Stretching vibrations: 3 and 4
Highly symmetric mode: 3
Number of bands expected to see in an experimental spectrum of CO2: 2 (Degeneracy means that 1 band is observed for 2 modes. Intensity of 3 are low)
| Wavenumber cm-1 | Symmetry | Intensity | |
|---|---|---|---|
| 1 | 640 | PIU | 145 |
| 2 | 640 | PIU | 13 |
| 3 | 1372 | SGG | 13 |
| 4 | 2436 | SGU | 1 |
Charge Analysis
Charge on C atom: 1.022
Charge on O atoms: -0.511
Since oxygen atoms are more electronegative than the carbon atom, negative charges are expected on the oxygen atoms and positive charge is expected on the carbon atom.
Molecular orbitals

Molecular orbital 4
Antibonding/bonding: Bonding
Symmetry: g
Occupied/unoccupied: Occupied
Contributing AOs: 2s(O), 2s(O)
Energy: -1.16099 a.u.
MO4 has a very high bonding character as no nodal plane is present, allowing the MO to take up the linear shape of the molecule. It is deep in energy and does not participate in chemical reactions. Carbon 2s AO is higher in energy than oxygen 2s AO and so it interacts with the oxygen 2p AOs instead, which is shown in MO6.
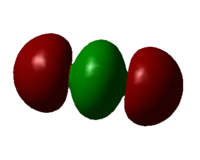
Molecular orbital 6
Antibonding/bonding: Bonding
Symmetry: g
Occupied/unoccupied: Occupied
Contributing AOs: 2p(O), 2s(C), 2p(O)
Energy: -0.56233 a.u.
The two 2p orbitals on each oxygen form sigma bonds with the centre carbon 2s orbital. This orbital is higher in energy than MO4 as there are 2 nodal planes. The large s character in the centre lobe shortens the bond length and increases the force constant of the bond.
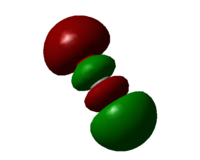
Molecular orbital 7
Antibonding/bonding: Bonding
Symmetry: u
Occupied/unoccupied: Occupied
Contributing AOs: 2p(O), 2p(C), 2p(O)
Energy: -0.51655 a.u.
Contrast with MO6, the 2p of carbon overlaps with the 2p of oxygen to form sigma bonds. The centre two lobes have antibonding character which will increase the bond length and decrease the force constance. There is now one more nodal plane and so the energy of MO7 is higher than MO6.
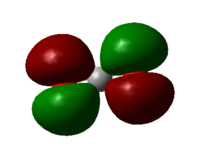
Molecular orbital 11
Antibonding/bonding: Antibonding
Symmetry: g
Occupied/unoccupied: Occupied
Contributing AOs: 2p(O), 2p(O)
Energy: -0.36997a.u.
HOMO of CO2, generated by the out of phase interaction of the 2p oxygen AOs. It is occupied by lone pairs and do not participate in bonding.
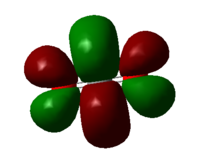
Molecular orbital 12
Antibonding/bonding: Antibonding
Symmetry: u
Occupied/unoccupied: Unoccupied
Contributing AOs: 2p(O), 2p(C), 2p(O)
Energy: 0.02992a.u.
LUMO of CO2, generated by the out of phase overlap of the 2p AOs on all the atoms. Each lobe repel each other which explains how the outer lobes are squeezed outwards.
PF3Cl2 molecule (Independence)
NPF3Cl2 molecule |
Calculation method: RB3LYP
Basis set: 6-31G(d,p)
Point group: C2v
Final energy E(RB3LYP): -1561.35287056 a.u.
RMS gradient: 0.00007156 a.u.
Optimised bond lengths:
| Axial / equatorial | Bond | Bond length A |
|---|---|---|
| Axial | P-F | 1.61 |
| Equitorial | P-F | 1.58 |
| Equatorial | P-Cl | 2.04 |
Optimised bond angles:
| Axial / equatorial | Bond | Bond angle ° |
|---|---|---|
| Axial | F-P-Cl | 90 |
| Axial | F-P-F | 89 |
| Equatorial | F-P-Cl | 119 |
| Equatorial | Cl-P-Cl | 122 |
Item Value Threshold Converged? Maximum Force 0.000134 0.000450 YES RMS Force 0.000046 0.000300 YES Maximum Displacement 0.001348 0.001800 YES RMS Displacement 0.000496 0.001200 YES
File:MMS4518 PF3CL2 OPTF POP.LOG
Vibrational Analysis
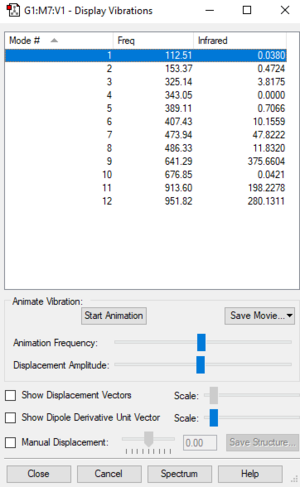
Number of modes expected from 3N-6 rule: 12
Degenerate modes: None
Bending vibrations: 1-8
Stretching vibrations: 9-12
Highly symmetric mode: 10
Number of bands expected to see in an experimental spectrum of PF3Cl2: 7 (Intensity of 1,2,4,5 and 10 are low)
| Wavenumber cm-1 | Symmetry | Intensity | |
|---|---|---|---|
| 1 | 113 | A1 | 0 |
| 2 | 153 | B1 | 0 |
| 3 | 325 | B2 | 4 |
| 4 | 343 | A2 | 0 |
| 5 | 389 | A1 | 1 |
| 6 | 407 | B1 | 10 |
| 7 | 474 | A1 | 48 |
| 8 | 486 | B2 | 12 |
| 9 | 641 | B1 | 376 |
| 10 | 677 | A1 | 0 |
| 11 | 914 | A1 | 198 |
| 12 | 952 | B2 | 280 |
Charge Analysis
| Axial / equatorial | Atom | Charge |
|---|---|---|
| Axial | F | -0.566 |
| Equatorial | F | -0.535 |
| Equatorial | Cl | -0.231 |
| -- | P | 2.128 |
Phosphorus is highly electropositive compared to the other atoms, so a positive charge is expected there. Negative charges are expected on the other atoms, of which fluorine will bear a more negative charge due to its higher electronegativity.
Reference
- ↑ Neal P. Mankad, Matthew T. Whited & Jonas C. Peters Prof. (2007) Terminal FeI-N2 and FeII⋅⋅⋅H-C Interactions Supported by Tris(phosphino)silyl Ligands. Angewandte Chemie International Edition 46 (30), 5768-5771. Available from DOI: 10.1002/anie.200701188
Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
NO
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES, you could have explained that the charges are 0 as the electronegativities are equal.
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
You have done a good job of presenting this information, well done! You mixed up the bonding nature of the MOs in some instances, e.g. you stated MO7 to be bonding but in the descriptive text you are quoting it to be anti-bonding. For MO 11 you are stating it to be antibonding and occupied but not having an effect on bonding.
Independence 1/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or
YES - well done!
Do some deeper analysis on your results so far
