Rep:Mod:MJM444
Starting optimisations
20140222 Saturday
Having drawn this taxol precursor in ChemDraw Pro 13.0 and optimised it to this structure with the MMFF94 force field in Avogadro, which I called the O-up isomer in my file names, I then pulled the atoms around while Avogadro's perpetual force field optimiser was running to produce this other isomer, which I called the O-down isomer in my file names. This too I optimised at with the MMFF94 force field. Moving straight on to try to generate some NMR spectra for the sulphur-containing derivatives 17 and 18, I added a hetrocyclic ring to each optimised structure by building in Avogadro's molecule editor, optimising both again at the MMFF94 level. I saved all my files as MOL files ready to import into GaussView to set up the more expensive calculations. Having a look at the paper with the spectroscopic references by Paqette, Pegg, Toops, Maynard, and Rogers (1990),[1] it seems that the geometry of the possible isomers is possibly fixed very nicely so that few confomers need be considered in finding the geometry of minimum energy: the fused ring systems have this 'rigid' property. The calculations might take some time nonetheless, so I first optimised the molecules to a minimum again, this time using the HF 3-21G model chemistry, not calculating force constants.
Once the first molecule was ready, I set up another optimisation, this time an optimsiation and frequency calculation, calculating the force constants once, at the B3LYP 6-31Gdp level, letting the calculation run for a few minutes to make sure that things looked okay before submitting it to high performance computing (HPC) to deal with. I submitted the second molecule in the same way, so that both 'O-up' and 'O-down' sets of files were going to soon be optimised first at the MMFF94 level, then at the HF 3-21G level and then at the B3LYP 6-31Gdp level ready for calculating the NMR spectra for the molecules.
An example of the lines at the top of the Gaussian input file is shown here for the 'O-down' isomer:
%chk=C:\G09W\Scratch\20140222 Taxol derivative O down B3LYP 6-31Gdp opt freq 1a.chk
# opt freq b3lyp/6-31g(d,p) geom=connectivity
These were left to run overnight.
Trying to calculate the NMR spectrum for H2salen, a precursor for salcomine I had synthesised in the lab a few weeks ago, I submitted an optimisation and frequency calculation to Gaussian overnight a few days ago but the calculation had failed. Fingers crossed...
Returning to check the optimisations
20140223 Sunday
Both calculations ran successfully overnight and perhaps, since it is the weekend, partly into the day. The 'O-up' isomer shown here had an energy of -1612.49376171 a.u. and its vibrational frequencies (shown below - click to enlarge the image) showed that the calculations had indeed found an energy minimum, since all the frequencies were real.

The 'O-down' isomer shown here had a slightly higher energy of -1612.48877961 a.u. suggesting that it would occupy a smaller portion of an equilibrium between the two isomers studied. Again, this isomer's vibrational frequencies (below) were all real, showing that the calculation indeed optimised to an energy minimum as it should do.

Submitting NMR calculations
The O-up isomer 17 has experimental prton NMR data run in deuterated chloroform.[1] Checking the boxes to set up the calculation in GaussView, and entering the keywords 'freq' and 'EmpericalDispersion=GD3' the command line in the file (below) shows that the calculation not seems to be at the 'rb3lyp/6-31g(d,p)' level rather than the 'b3lyp/6-31g(d,p)' level. The 'r' before the 'b3lyp/6-31g(d,p)' seems to stand for 'restricted'...
%chk=C:\G09W\Scratch\20140223 O-up NMR freq B3LYP 6-31Gdp 1a.chk
# nmr=giao rb3lyp/6-31g(d,p) scrf=(cpcm,solvent=chloroform) geom=connectivity freq empiricaldispersion=gd3
Running this on the PC for a while to confirm that the calculation runs for a few minutes without crashing, this was then submitted to the HPC suite. A calculation was set up, for interest (to see any differences), with a default spin but otherwise identical with the following command line, and submitted too.
%chk=C:\G09W\Scratch\20140223 O-up NMR freq B3LYP 6-31Gdp 1b default spin.chk
# nmr=giao b3lyp/6-31g(d,p) scrf=(cpcm,solvent=chloroform) geom=connectivity freq empiricaldispersion=gd3
The proton NMR spectrum of the O-down isomer 18 in the literature was run in deuterated benzene instead of deuterated chloroform.[1] Attempting to set up the calculation again in this solvent for this isomer proved initially difficult, with the calculation crashing within seconds when run initially on the PC. Closing the program and trying again, with the following command line, the calculation seemed to run just fine initially.
%chk=C:\G09W\Scratch\20140223 O-down NMR freq B3LYP 6-31Gdp 1a default spin.chk
# nmr=giao b3lyp/6-31g(d,p) scrf=(cpcm,solvent=benzene) geom=connectivity freq empiricaldispersion=gd3
Leaving the calculations to run overnight, with fingers crossed again, it would be interesting to know how optimising the molecules first speeds up the calculations. With that in mind, the molecules were not optmised at the restricted level, so I will delete this calculation from the list and run it again, for interest, if time allows: recalculating again at a different level of theory would not be quick and efficient! (Done - the deleted calculation was reported to have run for one hour and fifteen minutes, it appears.)
NMR spectra of the isomers
20140224 Monday
The NMR calculation seems to have worked well, and the predicted spectra are ready for inspection. The proton spectrum simulated in chloroform (with 'TMS B3LYP/6-31G(d,p) Chloroform' as the reference choice) is shown below for the 'O-up' isomer 17. (Click the scalable vector image to enlarge it.)

The predicted proton NMR spectrum for the 'O-down' isomer 18 is shown below (with 'TMS B3LYP/6-31G(d,p) Benzene' as the reference choice).

Considering the two spectra together, might it be possible to find evidence that Paqette, Pegg, Toops, Maynard, and Rogers (1990) have assigned the correct isomers, 17 and 18, to their corresponding spectra?
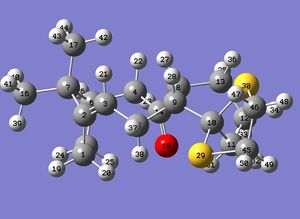
Looking at the numbering used in identifying the atoms in the molecule, it should be notes that the numbers are identical except that they go in exactly opposite directions around the sulphur-containing ring (click the numbering pictures below to enlarge). The numbering for isomer 17 is shown here:
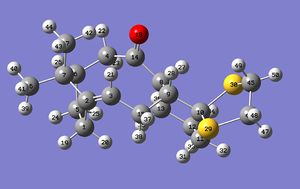
The numbering for 18 is shown here:
It appears that the program generating the NMR spectra is treating the molecule as rigid, with one proton (number 42) of one of the methyl groups much more deshielded (at 2.3 ppm) than the one nearest to the double bond (number 43 at 0.9 ppm). No conclusions possible yet...
Checking the frequencies of the molecules generated last night, while the 'O-down' isomer has only real vibratonal frequencies, confirming that the calculation has located an energy minimum, (the bottom of the potential-energy valley,) the 'O-up' isomer has one imaginary frequency, -31.10 /cm, suggesting that the program has found a transition state (a conformation that is an energy maximum not an energy minimum - the top of a hill where the potential energy gradient does not change with atom coordinates but is an unstable one nonetheless). The molecules were both possessed of only real frequencies yesterday - one of them must have re-optimised its geometry overnight when the additional NMR calculation was performed.
Averaging the chemical shifts for the rotating methyl groups
20140225 Tuesday
The molecules' geometries were calculated at a theoretical temperature of zero Kelvin. The molecules in the literature would have been investigated at around three-hundred Kelvin (room temperature). At this temperature, the methyl groups would be rotating, indistinguishable on the NMR timescale. Having been doing some thinking overnight, without any calculations running on the computers, it appears to me now that the predicted signals for the individual protons in the methyl groups could be averaged together. The weighted average should replicate the literature values.
Starting with the O-down isomer 18 (optimised correctly to an energy minimum), protons numbered 39, 40 and 41 belong to one freely-rotating methyl group. Taking the mean value of their respective predicted chemical shifts in benzene with a TMS reference, 1.497021,0.975694 and0.975694, gives their new predicted room temperature signal at 1.149469 ppm.
Protons numbered 42, 43 and 44 have chemical shifts at 1.32076,1.221034 and 1.097374 ppm respectively, with a mean value of 1.213056 ppm, their new room temperature predicted signal.
| Chemical shift /ppm | Atom number |
|---|---|
| 5.31 | 21 |
| 3.28 | 47 |
| 3.12 | 27, 48 |
| 2.99 | 28, 31, 49, 50 |
| 2.62 | 22, 37 |
| 2.31 | 25, 38 |
| 2.18 | 20 |
| 2.04 | 19, 26 |
| 1.91 | 24 |
| 1.77 | 23, 32, 34, 36 |
| 1.50 | 33, 35 |
| 1.21 | 39, 40, 41 |
| 1.15 | 42, 43, 44 |
Attempting to group these as in the literature, the predicted chemical shifts for 18 can be quoted (making no assumptions about the splitting of peaks that are not multiplets except for the fair assumption that the rotating methyl groups will be singlets) and compared:
1H NMR (predicted, C6D6) chemical shift 5.31 (1H), 3.28 (1H), 3.12-2.99 (m, 6H), 2.62-2.31 (m, 4H), 2.18-1.77 (m, 8H), 1.50 (2H), 1.21 (s, 3H, Me), 1.15 (s, 3H, Me),
(lit.[1], 1H NMR (300 MHz, C6D6) chemical shift 5.21 (m, 1H), 3.00-2.70 (m, 6H), 2.70-2.35 (m, 4H), 2.20-1.70 (m, 6H), 1.58 (t, J = 5.4 Hz, 1H), 1.50-1.20 (m, 3H), 1.10 (s, 3H), 1.07 (s, 3H), 1.03 (s, 3H)).
My molecule seems to have two protons missing... funny that...
Ooops! I have been modelling the wrong molecule! Isomer 18 has three methyl groups, whereas the molecule I have been modelling has a proton on the ring junction where a methyl group should be. Back to the drawing board!
Adding a methyl group
Starting over, or, at least, making a slight modification and recalculating, a proton on the ring junction was substituted for a methyl group on my old version of the 'O-up' isomer 18 to make the 'real' isomer 18; the molecule was optimised at the HF/3-21G level on the PC and sent to the HPC suite for NMR calculations at the B3LYP/6-31Gdp level skipping the optimisation at this level. Hopefully the molecule will be optimised to an energy minimum this time, with positive frequencies only. The top lines form the submitted job:
%chk=C:\G09W\Scratch\20140225 O-up 18 nmr opt freq gd3 chloro B3LYP 6-31Gdp.chk
'#' opt=calcfc freq b3lyp/6-31g(d,p) scrf=(cpcm,solvent=chloroform) geom=connectivity nmr empiricaldispersion=gd3
For 'O-down' isomer 17, also initially missing its methyl group on the ring junction, the methyl group was simply popped onto the ring junction in place of the hydrogen; this molecule is already optimised to an energy minimum at the B3LYP/6-31Gdp level so this may be quicker (it would be interesting to know the difference in the calculation times). The command line form this submitted job:
%chk=C:\G09W\Scratch\20140225 O-down 18 nmr opt freq gd3 chloro (once) B3LYP 6-31Gdp.chk
'#' opt=calcfc freq b3lyp/6-31g(d,p) scrf=(cpcm,solvent=benzene) geom=connectivity nmr empiricaldispersion=gd3
Proton NMR prediction for the corrected structures
20140226 Wednesday
Last night's calculations produced optimised molecules 17 and 18 with real vibrational frequencies only, as hoped. The further optimisation of (and NMR calculation for) the already HF/3-21G optimised structure 17 was fastest, taking two hours and thirty minutes to run. The optimisation and NMR calculation performed on structure 18, in which the methyl group was substituted onto the erroneous structure without optimisation at the HF/3-21G level, took longer, taking three hours and eight minutes to run. Although the difference is not huge, it appears that building up the optimisation in steps of increasing computational expense is faster overall than going straight in at the most expensive level (although identical confomers should really be compared for a truly fair analysis).
The corrected 'O-up' isomer
The optimised structure of the corrected 'O-up' isomer 17, is shown here and is shown in the picture below with atom numbering (right clicking on the link in this paragraph, and choosing Style > Labels > With Atom Number will display numbers on the atoms).

Averaging the chemical shifts for the freely rotating methyl groups including the newly added methyl group (protons 51, 52 and 53), the table below shows the predicted chemical shifts for isomer 17.
| Chemical shift /ppm | Proton number |
|---|---|
| 5.15 | 21 |
| 3.31 | 48 |
| 3.22 | 49 |
| 3.26 | 27 |
| 3.01 | 46, 47 |
| 2.72 | 20, 37 |
| 2.40 | 22, 23, 30, 36 |
| 2.27 | 19, 24 |
| 2.12 | 26, 34 |
| 1.95 | 31 |
| 1.90 | 33 |
| 1.60 | 25, 32 |
| 1.49 | 35 |
| 1.46 | 41, 42, 43 |
| 1.17 | 51, 52, 53 |
| 1.14 | 38, 39, 40 |
Listing in groups and comparing as before, it seems possible to make the data fit; methyl groups, all attached to carbons with no protons, are all assumed to be a 3H singlet:
1H NMR (predicted, CDCl3) chemical shift 5.15 (1H), 3.31-3.01 (m, 5H), 2.72-1.60 (m, 14H), 1.49 (1H), 1.46 (s, 3H, Me), 1.17 (s, 3H, Me), 1.14 (s, 3H, Me),
(lit.[1], 1H NMR (300 MHz, CDCl3) chemical shift 4.84 (dd, J = 7.2, 4.7 Hz, 1H), 3.40-3.10 (m, 4H), 2.99 (dd, J = 6.8, 5.2 Hz, 1H), 2.80-1.35 (series of m, 14H), 1.38 (s, 3H), 1.25 (s, 3H), 1.10 (s, 3H), 1.00-0.80 (m, 1H)).
The computer model predicts three methyl group signals that compare well with the 3H singlets seen experimentally, all predicted below 1.5 ppm and observed below 1.4 ppm experimentally. Also in the same region is a single proton signal, although its position is not predicted exactly. At the other end of the spectrum, there is a deshielded proton predicted at 5.15 ppm for the alkene proton. Attached to an alkene, this proton should be expected to be deshileded. There is a proton at 4.84 ppm experimentally. While the central group of fourteen proton shifts described in the literature agrees with the prediction, the doublet of doublets at 2.99 ppm adjacent to a group of four more deshilelded protons is not seen as a result of calculations; a group of five protons is noted instead. Nonetheless, considering that the B3LYP/6-31Gdp model chemistry is a compromise between accuracy and computational expense, the prediction appears to be reasonably close to experiment.
The corrected 'O-down' isomer
The optimised structure of the corrected 'O-down' isomer 18 shown here is shown in below with atom numbering.

Averaging the chemical shifts for the freely rotating methyl groups again, the table below shows the predicted chemical shifts for isomer 18.
| Chemical shift /ppm | Proton number |
|---|---|
| 5.46 | 21 |
| 3.22 | 46 |
| 3.09 | 47 |
| 2.92 | 48, 49 |
| 2.78 | 22 |
| 2.68 | 30 |
| 2.63 | 27 |
| 2.48 | 25 |
| 2.36 | 36, 37 |
| 2.19 | 20 |
| 2.00 | 26, 19 |
| 1.87 | 24 |
| 1.76 | 31, 32, 33 |
| 1.54 | 23, 35 |
| 1.44 | 51, 52, 53 |
| 1.27 | 34 |
| 1.24 | 41, 42, 43 |
| 1.16 | 38, 39, 40 |
Listing in matching groups where possible and comparing as before:
1H NMR (predicted, C6D6) chemical shift 5.46 (1H), 3.22-2.68 (m, 6H), 2.63-2.36 (m, 4H), 2.19-1.76 (m, 7H), 1.54 (2H), 1.44 (s, 3H, Me), 1.27 (1H), 1.24 (s, 3H, Me), 1.16 (s, 3H, Me),
(lit.[1], 1H NMR (300 MHz, C6D6) chemical shift 5.21 (m, 1H), 3.00-2.70 (m, 6H), 2.70-2.35 (m, 4H), 2.20-1.70 (m, 6H), 1.58 (t, J = 5.4 Hz, 1H), 1.50-1.20 (m, 3H), 1.10 (s, 3H), 1.07 (s, 3H), 1.03 (s, 3H)).
Three methyl 3H triplets and a 1H signal are predicted below 1.5 ppm, agreeing roughly with the literature signal patterns and shifts for both isomers 17 and 18. The chemical shift for a single proton observed experimentally at 5.21 ppm agrees with that calculated here for the 'O-down' isomer 18 (5.46 ppm) but the most deshielded proton predicted for the 'O-up' isomer 17 5.15 ppm also agrees with this. When the remaining chemical shifts are put into groups, the predicted pattern for 18 (1H, 6H, 4H, 7H, 2H, 3H, 1H, 3H, 3H) can be taken to fit the experimental pattern for 18 (1H, 6H, 4H, 6H, 1H, 3H, 3H, 3H, 3H) approximately. Compare the pattern predicted for 17 (1H, 5H, 14H, 1H, 3H, 3H, 3H) with the pattern reported experimentally for 18, and the approximate fit in this case too could leave doubts about the assignment of the spectra to the isomers. Note that the experimentally determined spectrum reported for isomer 17 (1H, 4H, 1H, 14H, 3H, 3H, 3H, 1H) could also be made to appear to fit the data predicted for isomer 18 with a little regrouping. Comparing the chemical shifts of the peaks, either of the predicted spectra could be taken to match either of the experimental proton spectra roughly equally well given the correct prediction of the alkene proton to be deshielded in both predictions and the methyl groups to be deshielded, with variation in both cases as to the location of the singlet nearest to the methyl groups in the spectrum. A look at the splitting pattern, with coupling constants reported in Hertz by the experimenters, could be useful in making spectral assignments.
Coupling constants
Two new jobs have been submitted to the 'Gaussian - 8px' application in the HPC suite: calculations of the spin-spin coupling constants for isomers 17 and 18. The command lines differ in the solvent used for the simulation, and are as follows. For 17:
%chk=C:\G09W\Scratch\20140226 O-up 17 nmr freq all-couplings B3LYP 6-31Gdp.chk
# nmr=(giao,spinspin,mixed) b3lyp/6-31g(d,p) scrf=(cpcm,solvent=chloroform) geom=connectivity freq empiricaldispersion=gd3
For 18:
%chk=C:\G09W\Scratch\20140226 O-down 18 nmr freq all-couplings benz B3LYP 6-31Gdp.chk
# nmr=(giao,spinspin,mixed) b3lyp/6-31g(d,p) scrf=(cpcm,solvent=benzene) geom=connectivity freq empiricaldispersion=gd3
Both calculations ran for a minute or two on the desktop computer without problems, so the predictions will hopefully be ready for inspection tomorrow. In the mean time, it would be interesting to get to know the program ChemBio3D.
Modelling cyclopentadiene dimers
This product of the dimerisation reaction of cyclopentadiene was drawn in ChemDraw Pro 13.0 and loaded in ChemBio3D Ultra 13.0. This cyclopenatdiene dimer with a trans ring junction was immediately displayed without any questions asked (Avogadro asks first if the structure should be made three-dimensional). Moving atoms and optimising several times at the MMFF94 level of theory produced this exo cycloaddition product 1 (with an energy of 55.3774 kcal/mol) and this endo reaction product 2 (with an energy of 58.1914 kcal/mol). While the process seemed simple enough, each optimisation resulted in a product with slightly lower energy than before. Avogadro will continue optimisation indefinitely, showing when the rate of change of potential energy has reached zero so that it is possible to know for sure when the energy minimum has been located. Preferring Avogadro's perpetual force optimiser, I will work with Avogadro from now. This will allow me to recalculate at the MMFF94s level. It appears at this stage that the exo isomer 1 is more stable, implying that this pericyclic reaction is under kinetic control (since the less thermodynamically stable endo isomer 2 is formed exclusively in reactions).
Optimising the endo isomer 2 in Avogadro (version 1.1.1) at the MMFF94s level of theory gave this structure of energy 58.19069 kcal/mol. The components contributing to the energy are shown in the table below.
| Energy type | Energy /(kcal/mol) |
|---|---|
| TOTAL BOND STRETCHING ENERGY | 3.46740 |
| TOTAL ANGLE BENDING ENERGY | 33.19104 |
| TOTAL STRETCH BENDING ENERGY | -2.08217 |
| TOTAL TORSIONAL ENERGY | -2.94985 |
| TOTAL OUT-OF-PLANE BENDING ENERGY | 0.02196 |
| TOTAL VAN DER WAALS ENERGY | 12.35756 |
| TOTAL ELECTROSTATIC ENERGY | 14.18475 |
| TOTAL ENERGY | 58.19069 |
Optimising the exo isomer 1 in Avogadro at the MMFF94s level of theory gave this structure of energy 55.37344 kcal/mol, agreeing with ChemBio3D that this isomer is lowest in energy. (Both approaches should agree: MMFF94 and MMFF94s model chemistries are both mechanical force fields.) Component energies are shown below.
| Energy type | Energy /(kcal/mol) |
|---|---|
| TOTAL BOND STRETCHING ENERGY | 3.54300 |
| TOTAL ANGLE BENDING ENERGY | 30.77270 |
| TOTAL STRETCH BENDING ENERGY | -2.04139 |
| TOTAL TORSIONAL ENERGY | -2.73103 |
| TOTAL OUT-OF-PLANE BENDING ENERGY | 0.01485 |
| TOTAL VAN DER WAALS ENERGY | 12.80163 |
| TOTAL ELECTROSTATIC ENERGY | 13.01367 |
| TOTAL ENERGY | 55.37344 |
Comparing the components of the total energy for the isomers, it appears that although the Van der Waals energy is slightly higher for the exo isomer 1, raising its energy, this is more than compensated for by the reduced angle bending energy and, to a lesser extent, electrostatic energy relative to the endo isomer 2. It appears that the exo isomer has less ring strain.
Modelling the hydrogenated cyclopentadiene dimers
Investigating the possible products from the hydrogenation of the endo cyclopentadiene dimer 2, note that there are two conformers of isomer 3. This conformer is the higher-energy conformer, with an energy of 50.72284 kcal/mol.
The table below shows the component energies of the lower energy (50.44568 kcal/mol) conformer of 3 shown here. This conformer is presumably lower in energy because the hydrogenated side of the molecule points away from the double bond.
| Energy type | Energy /(kcal/mol) |
|---|---|
| TOTAL BOND STRETCHING ENERGY | 3.31141 |
| TOTAL ANGLE BENDING ENERGY | 31.93584 |
| TOTAL STRETCH BENDING ENERGY | -2.10216 |
| TOTAL TORSIONAL ENERGY | -1.46979 |
| TOTAL OUT-OF-PLANE BENDING ENERGY | 0.01319 |
| TOTAL VAN DER WAALS ENERGY | 13.63767 |
| TOTAL ELECTROSTATIC ENERGY | 5.11951 |
| TOTAL ENERGY | 50.44568 |
The other hydrogenated isomer 4 has an energy of only 41.25749 kcal/mol. With no other conformers with lower energies that the simulation could find, this isomer would be the one expected to form under thermodynamic conditions. (This does not say anything, however, about the energetic level of the transition state and kinetics, so without this information, it could be assumed that this isomer 4 is indeed the one expected to be most likely to form.) For comparison, the components of the total energy of this molecule are shown in the table below.
| Energy type | Energy /(kcal/mol) |
|---|---|
| TOTAL BOND STRETCHING ENERGY | 2.82307 |
| TOTAL ANGLE BENDING ENERGY | 24.68545 |
| TOTAL STRETCH BENDING ENERGY | -1.65716 |
| TOTAL TORSIONAL ENERGY | -0.37821 |
| TOTAL OUT-OF-PLANE BENDING ENERGY | 0.00028 |
| TOTAL VAN DER WAALS ENERGY | 10.63703 |
| TOTAL ELECTROSTATIC ENERGY | 5.14702 |
| TOTAL ENERGY | 41.25749 |
Every component of the energy, except for the electrostatic energy, is lower. The release of ring strain in the bicyclic system seems to want to drive the hydrogenation reaction strongly.
Since both isomers 3 and 4 have energies lower than the starting endo dimer 2, under equilibriating conditions both would be expected to form to some degree (this assumes that there are no kinetic barriers to their formation). with low-energy isomer 4 forming a larger proportion of the product.
Carbon NMR and coupling constants for the taxol precursors
20140227 Thursday
Last night's calculations of the spin-spin coupling constants of 'O-up' isomer 17 and 'O-down' isomer 18 of the taxol synthetic precursor took 6 hours 17 minutes and 6 hours 14 minutes respectively on the 'Gaussian - 8px' application of the HPC suite - both really big calculations on structures that were already structurally optimised at the B3LYP/6-31Gdp level used for the calculations. Without knowing how to inspect the coupling constants just yet, the carbon NMR spectra will be examined below and compared to the literature findings.
Carbon NMR for the isomers
The predicted proton NMR spectrum in deuterated chloroform is shown below for isomer 17.

While some of the protons in this spectrum needed averaging due to their presence on freely rotating methyl groups, this does not apply to the carbon NMR predicted spectrum (in chloroform) shown below for the same isomer 17.

Carbon chemical shifts are listed in the table below.
| Chemical shift /ppm | Carbon number |
|---|---|
| 216.18 | 14 |
| 145.12 | 2 |
| 124.80 | 3 |
| 90.49 | 10 |
| 60.66 | 9 |
| 57.13 | 8 |
| 52.60 | 6 |
| 51.68 | 7 |
| 46.80 | 4 |
| 46.03 | 44 |
| 42.28 | 11 |
| 40.55 | 45 |
| 35.43 | 15 |
| 31.26 | 5 |
| 29.42 | 13 |
| 29.37 | 50 |
| 27.30 | 1 |
| 26.43 | 16 |
| 23.01 | 12 |
| 19.82 | 17 |
As should be expected, the unsaturated carbons are most deshielded, with the carbonyl carbon most deshielded of all followed by the alkene carbons.... but without further ado, this is the same pattern seen in both the other isomer, 18, and both literature molecules, so without being able to conclude very much from a carbon NMR analysis, the coupling constants will be looked at.
The predicted proton NMR spectrum in deuterated benzene is shown below for the 'O-down' isomer 18.

The predicted carbon NMR spectrum for the 'O-down' isomer 18 (in benzene) is shown here in graph form and tabulated:

| Chemical shift /ppm | Carbon number |
|---|---|
| 209.04 | 14 |
| 148.81 | 2 |
| 118.64 | 3 |
| 91.24 | 10 |
| 64.49 | 9 |
| 55.36 | 6 |
| 54.62 | 8 |
| 50.05 | 7 |
| 45.63 | 45 |
| 41.74 | 44 |
| 37.80 | 11 |
| 37.55 | 4 |
| 35.57 | 13 |
| 34.20 | 15 |
| 32.18 | 50 |
| 28.63 | 1 |
| 26.22 | 16 |
| 25.15 | 5 |
| 22.81 | 17 |
| 22.31 | 12 |
The patterns, while fitting the experimental data reasonably well, are both so similar that they should not be considered too deeply, although it should be noted that the range of chemical shifts agrees with the literature,[1] with the smaller range of shifts (from most deshielded to least deshielded) occuring for isomer 18 in both cases. This is positive evidence of the correct assignment.
Coupling constants for selected protons
Paqette, Pegg, Toops, Maynard, and Rogers (1990) reported several coupling constants for the proton NMR peaks.[1] The total nuclear spin-spin coupling, J (in Hz), is reported in the Gaussian log file for each pair of nuclei.
Assuming coupling between only the closest non-equivalent protons to be significant, the first peak to look at here will be the doublet of doublets seen at 4.84 ppm (J = 7.2, 4.7 Hz) for the 'O-up' isomer 17. This is the alkene proton, the most deshielded proton in the molecule, the peak predicted for proton number 21. This peak is reported at 5.21 ppm as a multiplet for the 'O-down' isomer 18, so there is a difference to investigate. This proton, in both isomers, couples with the nearest protons 19, 20, 36 and 37, all four bonds away. Protons numbered 19 and 20 could be predicted to give the greater coupling mignitudes because they couple through a double bond, with correspondingly higher s-core electron character in the bonds through which they communicate with proton 21.
For the 'O-up' isomer 17, the total nuclear spin-spin coupling J values shown in the log file are:
19 to 21: -0.321669D+01 Hz = -3.2 Hz
20 to 21: -0.158738D+01 Hz = -1.5 Hz
21 to 36: 0.558876D+01 Hz = 5.6 Hz
21 to 37: 0.126009D+02 Hz = 12.6 Hz
For the 'O-down' isomer 18, the total nuclear spin-spin coupling J values shown in the log file are:
19 to 21: -0.310729D+01 Hz = -3.1 Hz
20 to 21: -0.168527D+01 Hz = -1.7 Hz
21 to 36: 0.348312D+01 Hz = 3.5 Hz
21 to 37: 0.123011D+02 Hz = 12.3 Hz
Transition state analysis for the cyclodimerisation
It was assumed earlier that because the thermodynamically favourable isomer of the cyclodimerisation of cyclopentadiene is not formed, the reaction must be under kinetic control. Calculating the energies of the transition states should give conformation that the least thermodynamically stable endo isomer 2 has a lower energy transition state and should therefore be the kinetically favoured product.
Adapting the exo product 2 by drawing dotted lines to indicate 'half bonds' between the two cyclopentadiene halves of the molecule, and moving the two halves apart a little, this exo transition state was obtained. Optimised to a Berny transition state, calculating force constants once and using 'opt=noeigen' to stop the calculation terminating prematurely, this transition state was calculated at the HF/3-21G level in Gaussian. The energy of this structure is -383.38354481 a.u.. The transition state has one imaginary frequency, as expected for an energy maximum, at -719.17 /cm, corresponding, as shown in the picture below (clicking on the image will give a larger animated image) to the two halves coming together, just as required for this transition state.

Treating the endo product 1 in the same way, this HF/3-21G endo transition state was obtained. The structure has an energy of -383.38600018 a.u.. The imaginary frequency (only one as required for an energy maximum, -651.45 /cm) is animated below. It corresponds, correctly, to the cyclopentadiene monomers coming together.

The transition structure of dimer 1 is lower in energy than that of dimer 2 (by 0.00245537 a.u.) when using like-for-like computational calculations with the HF/3-21G model chemistry using the same program: the energies are directly comparable. This confirms that the endo product 1 will form more quickly, with less activation energy required for reaction then the thermodynamically more stable dimer 2. A look at the electron distribution in the molecule may give a clue about the stability of the transition state.
Molecular orbitals explain the reactivity
The endo transition state on the path to the formation of endo dimer 1 allows the close apposition of the reacting double bond systems, including the parts of these that do not directly react together to form bonds. The highest occupied molecular orbital (HOMO) can be seen to span between these double bonds in a bonding interacton (a single lobe with no nodes). The orbitals (for both transition states) are shown below. The models are interactive and can be manipulated (enlarged, rotated) by the reader.
Endo transition state highest occupied molecular orbital
Endo HOMO |
Exo transition state highest occupied molecular orbital
Exo HOMO |
The transition state belonging to the exo dimer 2 does not have supportive bonding interactions between the forming double bonds of the system, because they are far apart. These orbitals are higher in energy, illustrating graphically why the endo product 2 forms more quickly through the more stable of the two transition states.
