Rep:Mod:M4H1M4
NH₃ Molecule
| Calculation method | Basis set | RMS gradient (au) | Final Energy E (au) | Point Group | |
|---|---|---|---|---|---|
| Ammonia | B3LYP | 6-31G(d,p) | 0.00000485 | -56.55776873 | Cv3 |
NH₃ molecule |
NH bond distance = 1.02 Å
H-N-H Bond angle = 106 °
distances and angles should be considered accurate to ≈ 0.01Å and bond angles are accurate to ≈ 1°
Item table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000070 0.001800 YES RMS Displacement 0.000033 0.001200 YES

The optimisation file is liked to here
| Mode | 1 | 2 | 3 | 4 | 5 | 6 |
| Wavelength (cm^-1) | 1089 | 1694 | 1694 | 3461 | 3590 | 3590 |
| Symmetry | A1 | E | E | A1 | E | E |
| Intensity (Arbitrary Units) | 145 | 14 | 14 | 1 | 0.27 | 0.27 |
We expect to see 6 modes with 2 and 3 being degenerate as well as 5 and 6. 1,2 and 3 are bending vibrations and 4,5 and 6 are bond stretch vibrations. Mode 4 is highly symmetrical. Mode 1 is known as the umbrella mode. There is no dipole change in modes 5 and 6. Mode 2 and 3 will produce the same band and mode 1 will also produce a band. Therefore we should expect to see 2 bands in an experimental spectrum of gaseous ammonia.
In ammonia, nitrogen has a charge of -1.125 and hydrogen has a charge of 0.375. Nitrogen is more electronegative than hydrogen due to its higher nuclear charge. This causes nitrogen to attract the electron density from the hydrogen atoms resulting in the hydrogen becoming positively charged and itself negatively charged.
N₂ Molecule
| Calculation method | Basis set | RMS gradient (au) | Final Energy E (au) | Point Group | |
|---|---|---|---|---|---|
| Nitrogen | B3LYP | 6-31G(d,p) | 0.00247261 | -109.52412298 | D∞H |
N₂ molecule |
N≡N bond distance = 1.10 Å
Item table
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000006 0.000300 YES Maximum Displacement 0.000002 0.001800 YES RMS Displacement 0.000003 0.001200 YES

The optimisation file is liked to here
| Mode | 1 |
|---|---|
| Wavelength (cm^-1) | 2457 |
| Symmetry | SGG |
| Intensity (Arbitrary Units) | 0 |
Both nitrogen have a charge of 0 as the molecule is non-polar. Both atoms are the same so have the same electronegativity resulting in a net charge of 0.
H₂ Molecule
| Calculation method | Basis set | RMS gradient (au) | Final Energy E (au) | Point Group | |
|---|---|---|---|---|---|
| Hydrogen | B3LYP | 6-31G(d,p) | 0.09719500 | -1.15928020 | D∞H |
H₂ molecule |
H-H bond distance = 0.60 Å
Item table
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
The optimisation file is liked to here
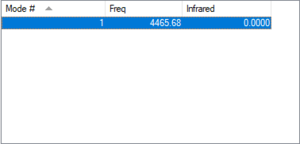
| Mode | 1 |
|---|---|
| Wavelength (cm^-1) | 4457 |
| Symmetry | SGG |
| Intensity (Arbitrary Units) | 0 |
The molecule is non-polar. As both atoms are the same they have a charge of 0.
Structure and reactivity
Comparing H-H bond in mono-metallic TM complex, ZUXSUG to H-H in hydrogen:
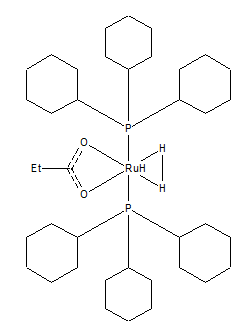
| H-H bond distance (Å) | |
| H₂ | 0.6 |
| ZUXSUG | 1.3 |
The H-H bond distance of the complex is over double that of my optimised H₂ molecule. This is due to the fact when hydrogen is attached to the more negative ruthenium ion the charge on the hydrogen also increases. This increase H-H repulsion increasing the bond angle and therefore the bond distance. There are no external factors such as transition metals affect the optimised H₂. The crystal structure and computational distance differ due to errors associated with calculating the values for each method.
Haber-Bosch Process Calculation
- E(NH3)= -56.5577687 au
- 2*E(NH3)= -113.1155370 au
- E(N2)= -109.52412868 au
- E(H2)= -1.1785394 au
- 3*E(H2)= -3.5356182 au
- ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.055791 au
Energy for reaction of N2 + 3H2 -> 2NH3 = -0.05580 au (-146.5 kJ/mol)
Therefore, as energy is released the forward process is favoured thermodynamically so ammonia is more stable than its substituents.
Small molecule: SiH₄
| Calculation method | Basis set | RMS gradient (au) | Final Energy E (au) | Point Group | |
|---|---|---|---|---|---|
| Silane | B3LYP | 6-31G(d,p) | 0.00005848 | -291.88802747 | TD |
SiH4 molecule |
Si-H bond distance = 1.48 Å
H-Si-H bond angle = 109 °
The literature value for the Si-H bond distance is 1.48 Å and 109.5° for the H-Si-H bond angle. These values are very similar or correct to 2 and 1 decimal places respectively.[1]
Item table
Item Value Threshold Converged? Maximum Force 0.000113 0.000450 YES RMS Force 0.000061 0.000300 YES Maximum Displacement 0.000589 0.001800 YES RMS Displacement 0.000315 0.001200 YES
The optimisation file is liked to here
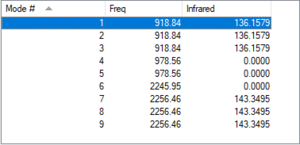
Using the 3N-6 rule we should expect to see 9 modes.
| Mode | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
| Wavelength (cm^-1) | 919 | 919 | 919 | 979 | 979 | 2246 | 2256 | 2256 | 2256 |
| Symmetry | TS2 | TS2 | TS2 | E | E | A1 | TS2 | TS2 | TS2 |
| Intensity (Arbitrary Units) | 136 | 136 | 136 | 0 | 0 | 0 | 143 | 143 | 143 |
Modes 1-5 are bending vibrations and 6-9 are stretching vibrations. Mode 6 is highly symmetrical. Modes 1,2 and 3 are degenerate as well as 4 and 5, and 7,8 and 9. therefore we should expect to see 3 bands in an experimental spectrum of silane.
The charge of the silicon atom is 0.630 and the charge of the hydrogen atoms are -0.157. Silicon is less electronegative than hydrogen as it is further down the periodic table so there is a larger distance between the nucleus and electron pairs, weakening the electrostatic attraction so electron density transfers to the hydrogen. Thereby leaving silicon with a positive charge and hydrogen with negative charge.

Energy calculations:
Si + 2H2 → SiH4
- E(SiH4)= -291.8880275 au
- E(Si)= -289.3289016 au
- E(H2)= -1.1592802 au
- 2*E(H2)= -2.3185604 au
- ΔE=E(SiH4)-[E(Si)+2*E(H2)]= -0.2405656 au

Energy of reaction = -0.24057 au = -631.6 kJ/mol therefore reaction is spontaneous.
Molecular orbitals



The symmetry of the 3s orbital is A₁ and the symmetry of the 3p orbitals are T₂.
This gives a bond order of 4.
The 1s orbital is not involved in chemical bonding as shown by the lack of overlap with the atomic orbitals of the hydrogen atoms and the low energy.
The 2s orbital is much higher in energy compared to the 1s orbital with an energy of -5.28 au, compared to the -66.13 au of the 1s orbital.
The 3s orbital is hybridised with three 3p orbitals to give an sp3 hybridised molecule, hence the tetrahedral symmetry. .A1 is formed combining 3s orbital with one of the 1s orbitals of the hydrogen atom. The overlap is more apparent here compared to the two previous MOs.
The MO with the T₂ symmetry is formed combining the three 3p orbitals with 3 of the hydrogen 1s orbitals, creating 3 degenerate levels, hence is higher up in energy(-0.3185 au) than MO 6 (-0.547 au). All pAO based MOs are occupied resulting in the high stability of Silane.
The 4s orbital is unoccupied and has a positive energy. The LUMO is closer to the 3p orbitals of the silicon and therefore SiH4 will act as an electron donor through the H atoms and electron acceptor through the silicon.
H₂O molecule
Decomposition of water 2H₂O → 2H₂ + O₂
E(H₂O) = -76.4197374 au
E(H₂) = -1.1785394 au
E(O₂) = -150.2574243 au
2*E(H₂O) = -152.8394750 au
2*E(H₂) = -2.3570788 au
ΔE=[E(O₂)+2*E(H2)]-2*E(H₂O) = 0.22497 au
=+590.7 kJ/mol
Reaction is endothermic therefore is unlikely to occur at room temperature.
References
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES - good structure well done!
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES - good explanations well done!
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
NO - you have included the identifier but not a link.
Have you compared your optimised bond distance to the crystal structure bond distance?
YES - you could explain more correctly on terms of back bonding, the coordination of H2 to Ru causes electron density to be added to the LUMO of the H2 which is an antibonding MO, thus reducing the bonding between the H atoms.
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 3/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES - good explanation about the charges.
You have some mistakes in your MO analysis - for example MO 6 is simply the interaction of the Si 3s AO with the 1s AOs on the H atoms, where all 5 AOs are the same phase. See the diagram included for how the MOs are formed from fragment MO diagrams of H2, then H4, then SiH4.

Independence 1/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far
YES you did some more energy calculations, well done!
