Rep:Mod:LWY0325
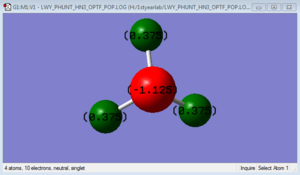
NH3 molecule
N-H bond length: 1.01865 Å H-N-H bond angle: 105.741 °
| Parameter | Data |
|---|---|
| calculation method | RB3LYP |
| basis set | 6-31G(d,p) |
| final energy E | -56.55776873 au |
| RMS gradient | 0.00000485 |
| point group | C3V |
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
test molecule |
The optimisation is link to the file: Media:LWY_PHUNT_HN3_OPTF_POP.LOG
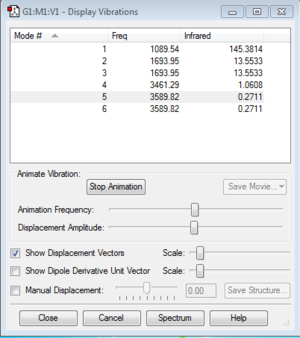
Display Vibrations
How many modes do you expect?
For non-linear molecule, the rule used to calculate the number of vibration modes is 3N-6, and N is the number of atoms in the molecule. This molecule has 4 atoms, so there are six vibration modes.
Which modes are degenerate (ie have the same energy)?
2&3, 5&6
Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Mode 1, 2, and 3 are bending vibrations; Mode 4, 5, and 6 are stretching vibrations.
Which mode is highly symmetric?
Mode 4
One mode is known as the "umbrella" mode, which one is this?
Mode 1
How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
We expect two bands on the infrared spectrum, although there are four distinguish frequencies for the vibration modes. The vibration modes 4, 5, and 6 have very low intensities, so we could not observe them on the spectrum. The modes 2 and 3 have the same frequency, so they contribute to a single peak.
Atomic Charges
The charge distribution on each atom depends on the electronegativity of that atom. The electronegativity of N is higher than H. This molecule is polar because of the difference of electronegativity between N and H, and N has a pair of electrons.
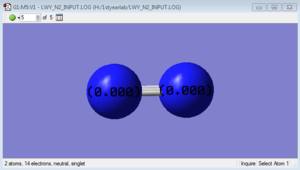
N2 molecule
N-N bond length: 1.09200 Å
| Parameter | Data |
|---|---|
| calculation method | RB3LYP |
| basis set | 6-31G(d,p) |
| final energy E | -109.52412868 au |
| RMS gradient | 0.00000365 |
| point group | D*H |
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000006 0.000300 YES Maximum Displacement 0.000002 0.001800 YES RMS Displacement 0.000003 0.001200 YES
test molecule |
The optimisation is link to the file: Media:LWY_N2_INPUT.LOG
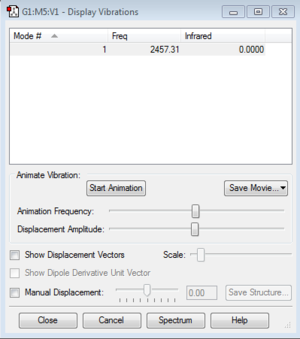
Display Vibrations
How many modes do you expect?
This is a linear molecule, so the rule used to calculate the number of vibration modes is 3N-5. There are two atoms in the molecule, so there is only one mode.
How many bands would you expect to see in an experimental spectrum?
There is no infrared band for N2, because the only vibration mode is streching, so there is no change in dipole in during the vibration.
Atomic Charges
The charge distribution is zero. This molecule is non-polar, and the two N have the same electronegativity.
H2 molecule
H-H bond length: 0.74289 Å
| Parameter | Data |
|---|---|
| calculation method | RB3LYP |
| basis set | 6-31G(d,p) |
| final energy E | -1.17853935 au |
| RMS gradient | 0.00003809 |
| point group | D*H |
Item Value Threshold Converged? Maximum Force 0.000066 0.000450 YES RMS Force 0.000066 0.000300 YES Maximum Displacement 0.000087 0.001800 YES RMS Displacement 0.000123 0.001200 YES
test molecule |
The optimisation is link to the file: Media:LWY_H2_INPUT.LOG
Display Vibrations
How many modes do you expect?
This is a linear molecule, so the rule used to calculate the number of vibration modes is 3N-5. There are two atoms in the molecule, so there is only one mode.
How many bands would you expect to see in an experimental spectrum?
There is no infrared band for H2, because the only vibration mode is streching, so there is no change in dipole in during the vibration.
Atomic Charges
The charge distribution is zero. This molecule is non-polar, and the two H have the same electronegativity, so there is no dipole in the molecule.
Reaction energy of N2 + 3H2 -> 2NH3
•E(NH3)= -56.55776873 au •2*E(NH3)= -113.11553746 au •E(N2)= -109.52412868 au •E(H2)= -1.17853935 au •3*E(H2)= -3.53561805 au •ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= 0.05579073 au (Converted to -146.478561615 kJ/mol) •The product energy is lower than that of the reactants, so the difference in energy is negative. The ammonia product is more stable.
Cl2 molecule
Cl-Cl bond length: 2.04163 Å
| Parameter | Data |
|---|---|
| calculation method | RB3LYP |
| basis set | 6-31G(d,p) |
| final energy E | -920.34987887 au |
| RMS gradient | 0.00000380 |
| point group | D*H |
Item Value Threshold Converged? Maximum Force 0.000007 0.000450 YES RMS Force 0.000007 0.000300 YES Maximum Displacement 0.000018 0.001800 YES RMS Displacement 0.000026 0.001200 YES
test molecule |
The optimisation is link to the file: Media:LWY_CL2_INPUT_OPTIMISE.LOG
Display Vibrations
How many modes do you expect?
This is a linear molecule, so the rule used to calculate the number of vibration modes is 3N-5. There are two atoms in the molecule, so there is only one mode.
How many bands would you expect to see in an experimental spectrum?
There is no infrared band for Cl2, because the only vibration mode is streching, so there is no change in dipole in during the vibration.
Atomic Charges
The charge distribution is zero. This molecule is non-polar, and the two Cl have the same electronegativity. There is no overall dipole on this molecule.
Molecular Orbitals
3s bonding orbital(occupied orbital)
3s AOs from the Cl atoms contribute to the 3s sigma MO bonding orbital. The energy related to the MO is -0.93315 au, which is much higher than the lowest MO. The energy of the lowest MO is -101.60297 au; the two core AOs contribute to this value. These low lying AOs hardly overlap, so the energy is very low. 3s MO is the first that contribute to overlap and bonding.
3s antibonding orbital(unoccupied orbital)
The energy of 3s antibonding orbital is -0.77745 au. 3s antibonding orbital has higher energy than the corresponding 3s bonding orbital, because it involves destabilisation. Moved from core electrons to valence electrons, the energy between the bonding orbital and the antibonding orbitals starts to increases, because the AOs in the valence shells overlaps more efficient. The antibonding MO decreases the bonding character of the molecule.
3p sigma bonding orbital(occupied orbital)
3p sigma bonding orbital has the energy of -0.47393 au. The two pAOs combined horizontally along the axis. This MO contributes to bonding.
3p pi bonding orbital(occupied orbital)
3p pi bonding orbital has the energy of -0.40696 au. Two pAOs bond parallel to each other and perpendicular to the horizontal axis, so the electron density is above and below the bond axis. There are two degenerate 3p pi bonding orbitals, which have higher energy than the 3p sigma orbital. This MO contributes to bonding.
3p pi antibonding orbital(unoccupied orbital)
The energy of the 3p pi antibonding orbital is -0.31360 au, and it is the MO that has the highest energy, so this MO is HOMO.The antibonding MO decreases the bonding character of the molecule.