Rep:Mod:KQ1623
NH3 molecule
Information about optimisation
NH3 bond distance=1.01798
NH3 bond angle=105.74115
Calculation Method = RB3LYP
Basis Set = 6-31G(d,p)
E(RB3LYP) = -56.55776873 a.u.
RMS Gradient Norm = 0.00000485 a.u.
Point Group = C3V
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
[here]
test molecule |
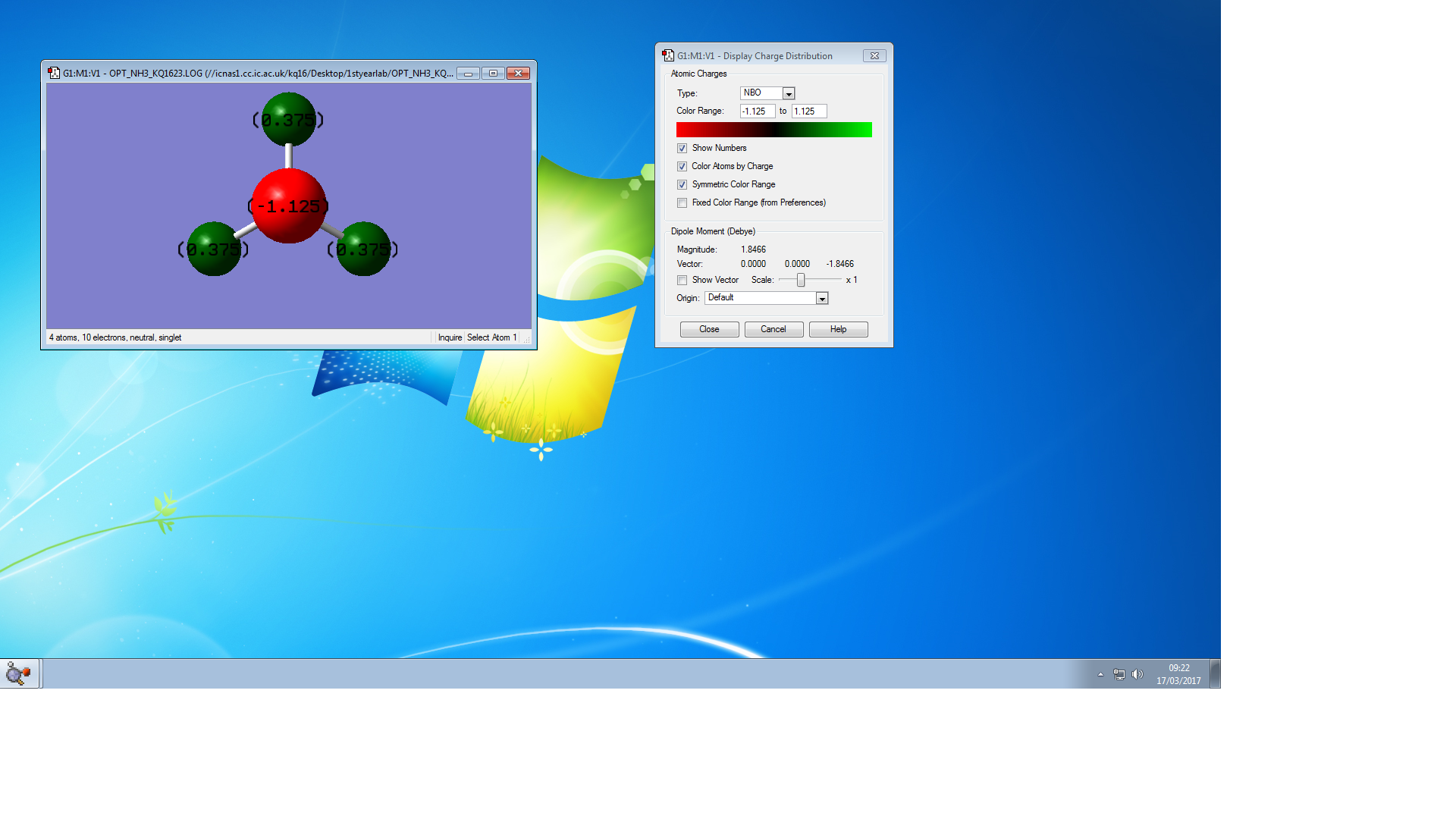
Charged Molecules
The charge on N atom is -1.125, and the charge of H atom is 0.37, I think that N should be negative because N is more electronegative
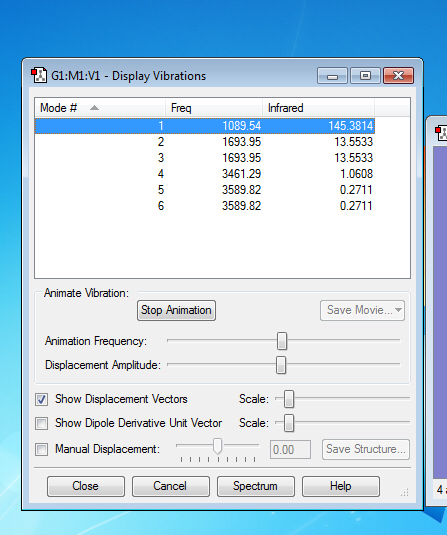
Vibrations
how many modes do you expect from the 3N-6 rule? 6
which modes are degenerate (ie have the same energy)? mode2 and 3, mode5 and 6
which modes are "bending" vibrations mode1,2 and 2 , which are "bond stretch" vibrations? mode4,5 and 6
which mode is highly symmetric? mode 1 4 6
one mode is known as the "umbrella" mode, which one is this? mode 1
how many bands would you expect to see in an experimental spectrum of gaseous ammonia? 2
Optimisation of N2
Bond length of N2= 1.10550
Calculation Method RB3LYP
Basis Set 6-31G(d,p)
E(RB3LYP) -109.52412868 a.u.
RMS Gradient Norm 0.00000060 a.u.
Point Group D*H
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401055D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
test molecule |
[here]
Optimisation of H2
Bond length of H2= 0.600000
Calculation Method RB3LYP
Basis Set 6-31G(d,p)
E(RB3LYP) -1.15928020 a.u.
RMS Gradient Norm 0.09719500 a.u.
Point Group D*H
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
test molecule |
[here]
Reaction Energies
E(NH3)= -56.55776873 a.u.
2*E(NH3)= -113.11553746 a.u.
E(N2)= -109.52412868 a.u.
E(H2)= -1.15928020 a.u.
3*E(H2)= -3.4778406 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.11356818
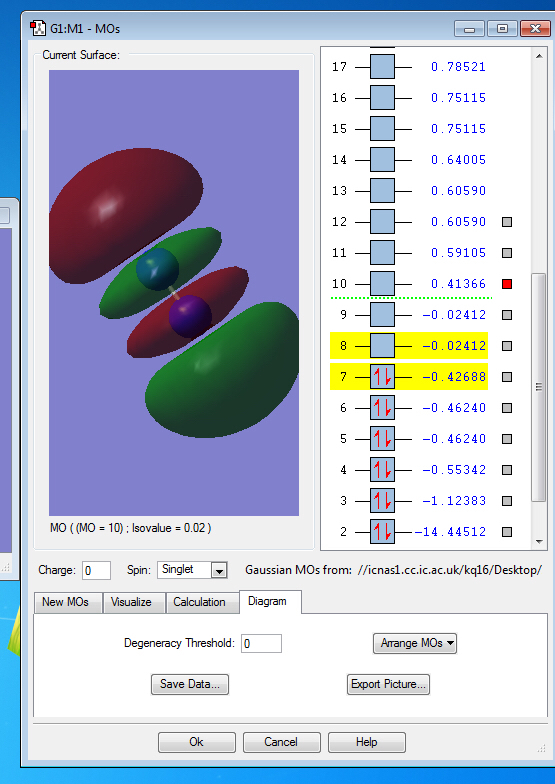
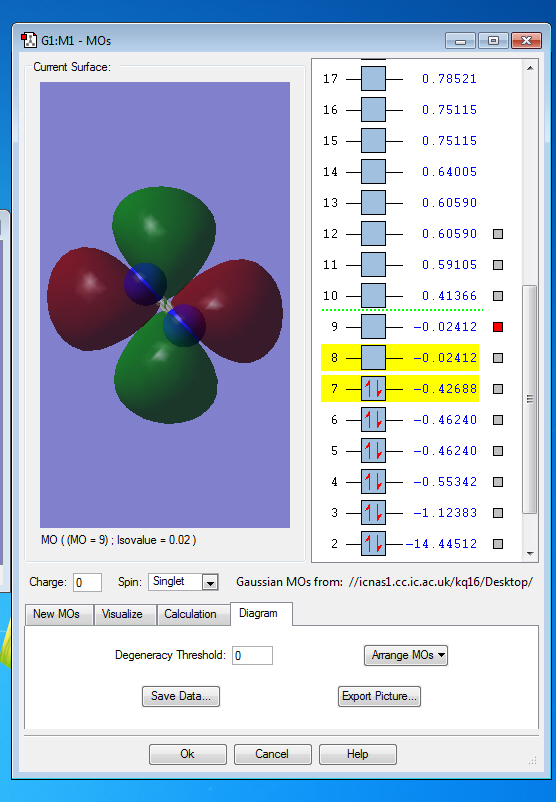
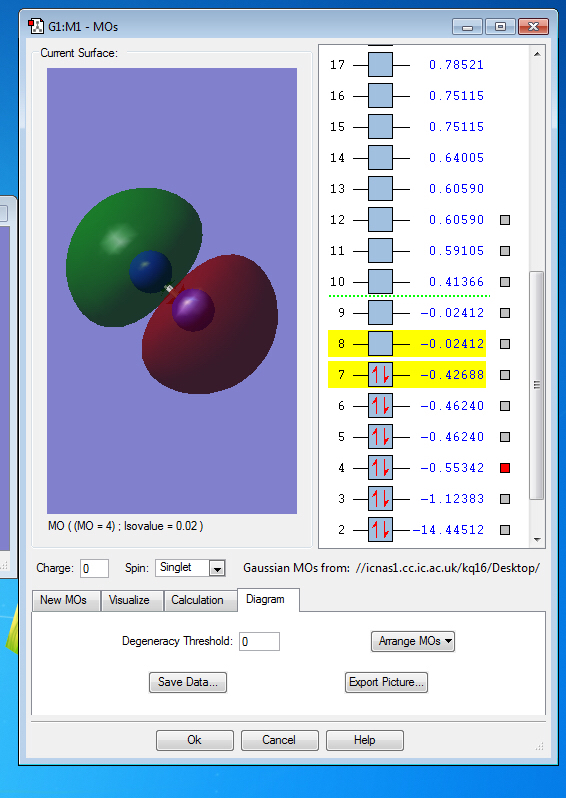
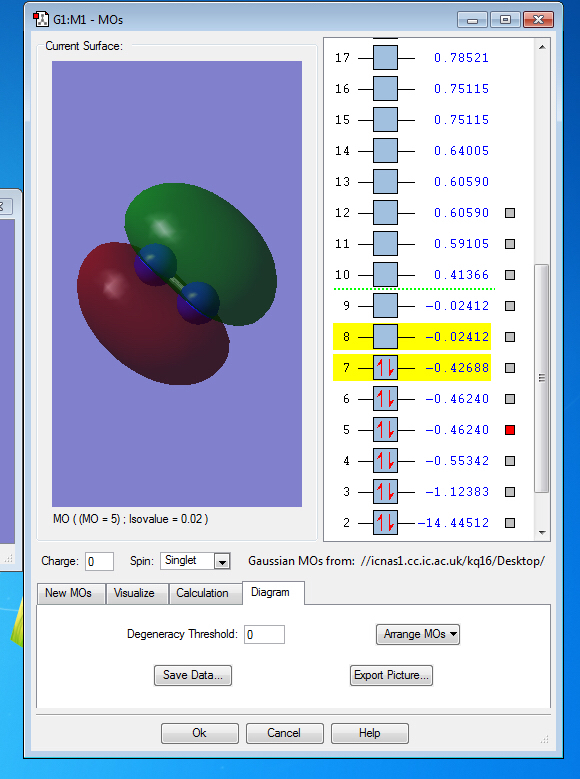
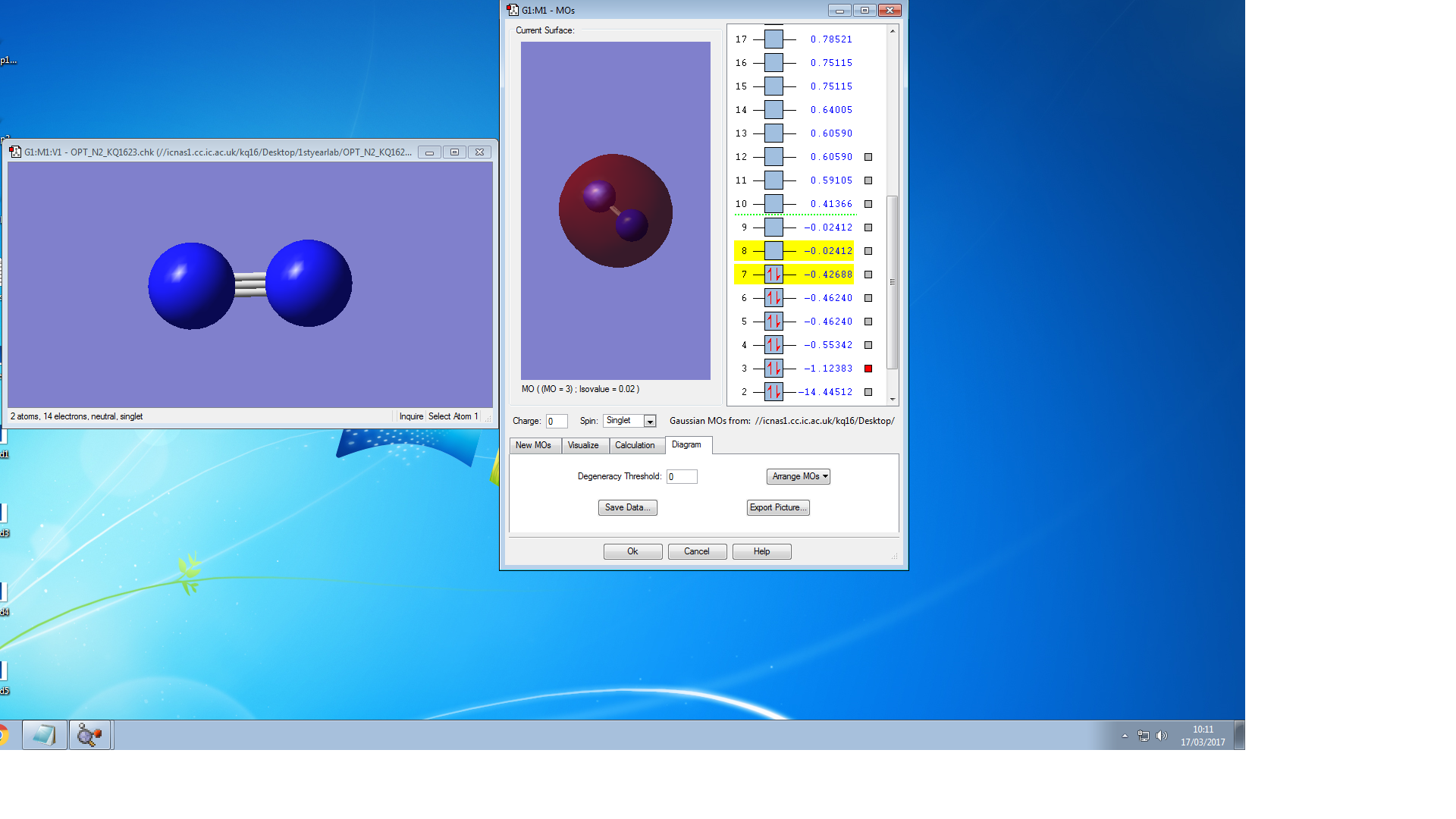
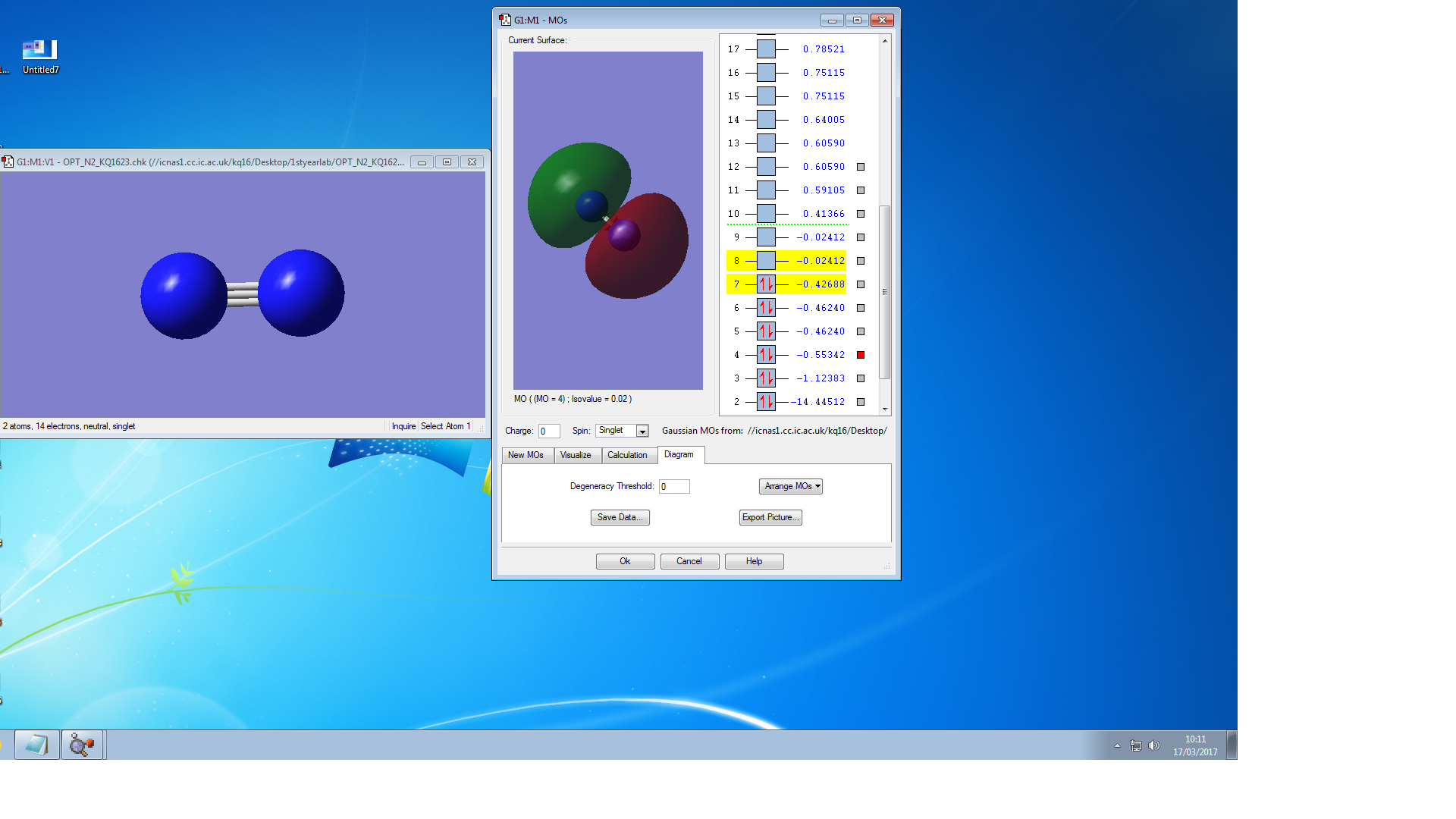
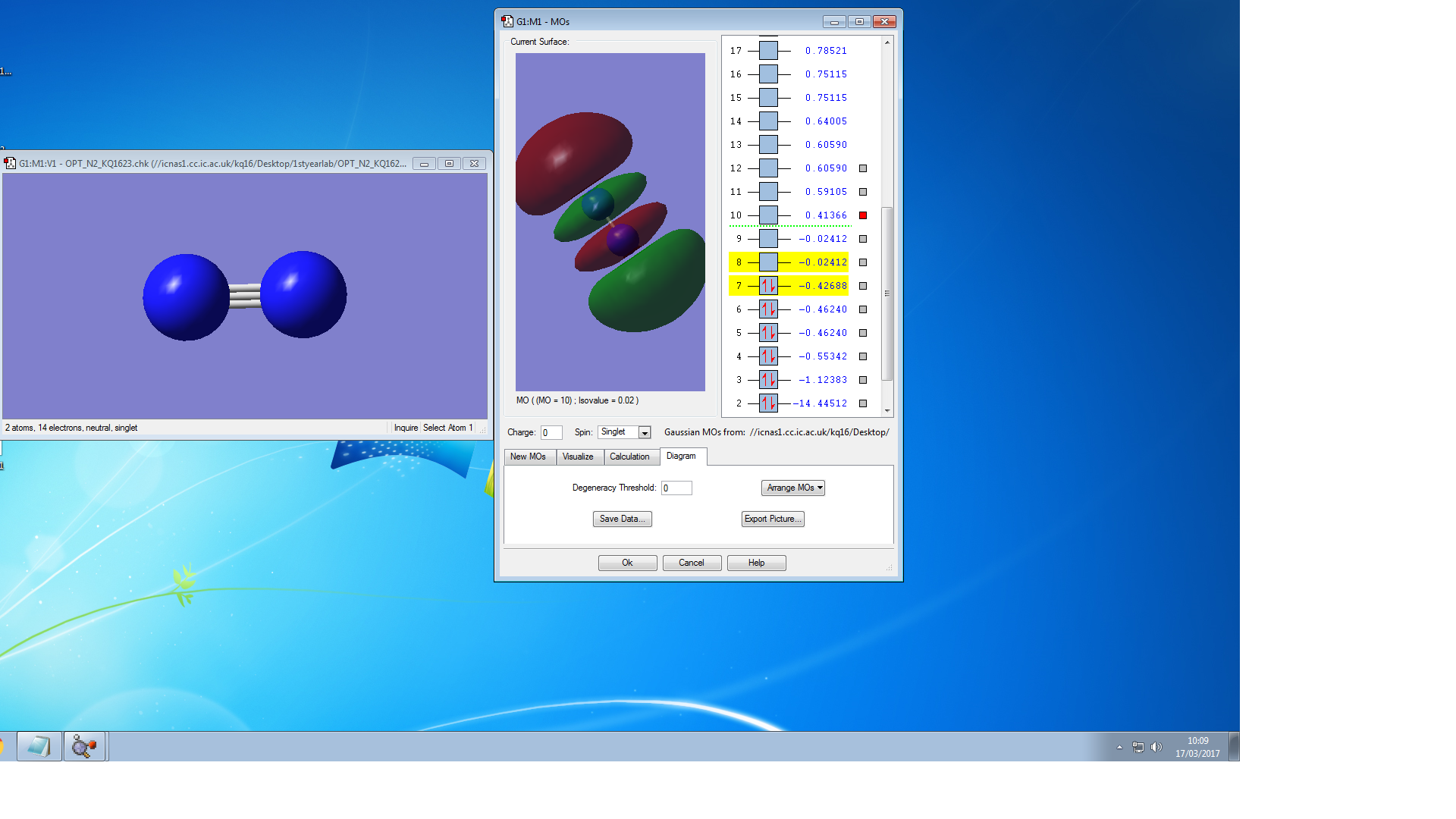
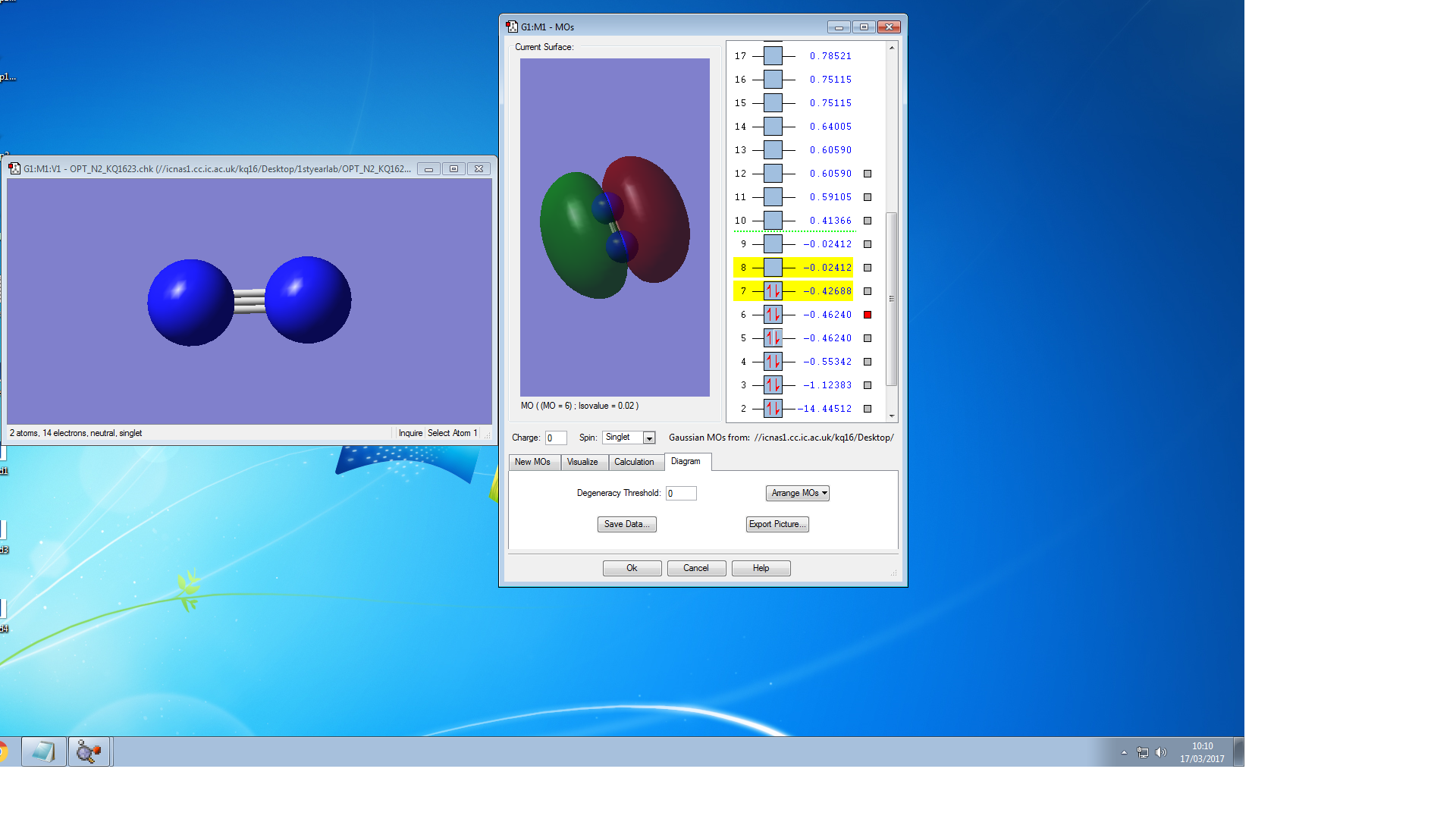
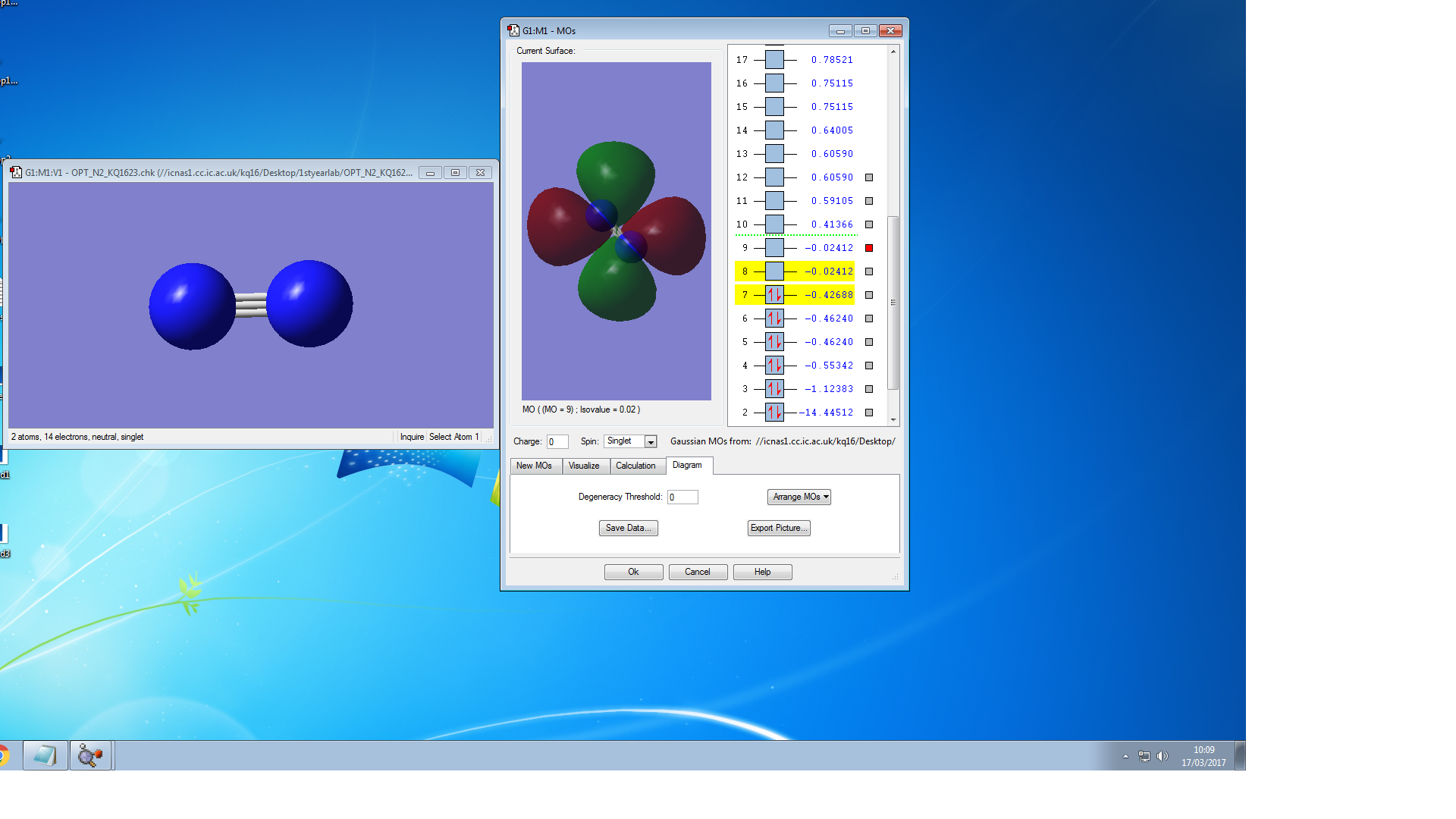
Molecular Orbitals of N2
F2 molecule
Information about optimisation
Bond length of F-F=1.16
Calculation Method RB3LYP
Basis Set 6-31G(d,p)
E(RB3LYP) -199.42620785 a.u.
RMS Gradient Norm 0.23253407 a.u.
Point Group D*H
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000128 0.000300 YES
Maximum Displacement 0.000156 0.001800 YES
RMS Displacement 0.000221 0.001200 YES
Predicted change in Energy=-1.995025D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4028 -DE/DX = 0.0001 !
--------------------------------------------------------------------------------
test molecule |
[[1]]
Vibrations
there is only one mode, it is bond stretch vibration
Molecular Orbitals
the symmetry of this MO is sigma
the symmetry of this MO is sigma*,it is occupied
It is unoccupied sigma MO
the symmetry of this MO is Pi, it is occupied
the symmetry of this MO is Pi*, it is unoccupied