Rep:Mod:KAT88811
Molecular modeling 2
NH3 molecule
Summary
•molecule name: Ammonia
•calculation method: B3LYP
•basis set: 6-31G(d,p)
•final energy E(RB3LYP) in atomic units (au): -56.55776873
•RMS gradientː 0.00000485
•the point group of your molecule: C3V
Bond length (A) = 1.01798
Bond angle (degrees) = 105.741
Item table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES Predicted change in Energy=-5.986281D-10
Molecular model
Optimized Ammonia |
My NH3 molecule link is here
Vibration modes
•how many modes do you expect from the 3N-6 rule?
(3x4) - 6 = 6 modes are expected
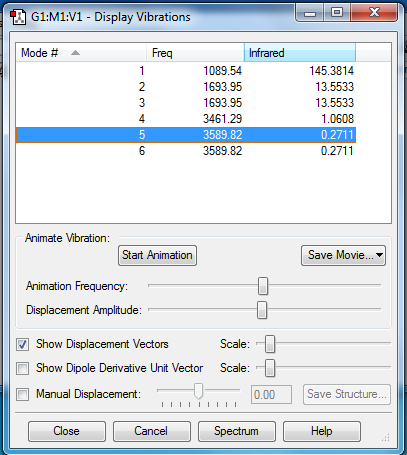
•which modes are degenerate (ie have the same energy)?
Modes 2-3 and 5-6 are degenerate
•which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending vibrations are modes 1-3; occur at lower frequencies as require less energy for motion. Bond stretching vibrations are modes 4-6; occur at higher frequencies as require more energy for initiation.
•which mode is highly symmetric?
Mode 1 is a symmetric bend, and mode 4 is a symmetric stretch.
•one mode is known as the "umbrella" mode, which one is this?
Mode 1
•how many bands would you expect to see in an experimental spectrum of gaseous ammonia?
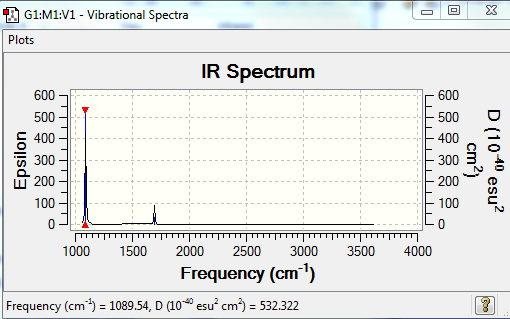
For a band/vibration to be seen on the IR spectrum a change in dipole should be observed. Therefore only two band will be seenː One for mode 1 with the greatest intensity of 145.3812 (Frequency 1089.54) and thus greatest movement of molecule along the dipole, and another one for modes 2 and 3 combined as they are degenerate - have the same IR intensity of 13.5533 (Frequency 1693.95).
Predicted IR spectrum of Ammonia
Charges
Charge on Nitrogen atom = -1.125 Charge on Hydrogen atom = 0.375
The electronegativity of N = 3.04 and H = 2.20 (Pauling scale). Therefore N is more electronegative than H thus attracts the shared electrons stronger to its nucleus, therefore N is expected to have a negative charge, and H is expected to have a positive charge. The calculated results support the theory stated.
H2 molecule
Summary
•molecule name: Dihydrogen/Molecular Hydrogen
•calculation method: B3LYP
•basis set: 6-31G(d,p)
•final energy E(RB3LYP) in atomic units (au): -1.17853936
•RMS gradientː 0.00000017
•the point group of your molecule: D∞h
Bond length (A) = 0.74279
Bond angle (degrees) = 180 (Linear molecule)
Item table
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES Predicted change in Energy=-1.164080D-13
Molecular model
Optimized Hydrogen molecule |
My H2 molecule link is here
Vibration modes
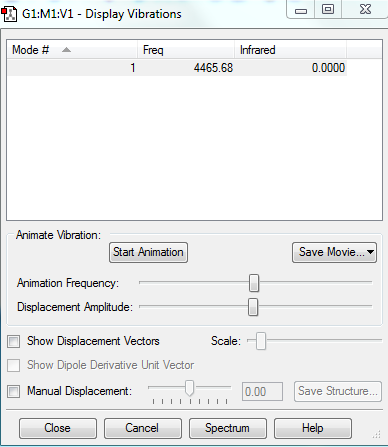
According to (3N - 5) rule only (6-5)= 1 mode of vibration expected, which is confirmed by the model computed.
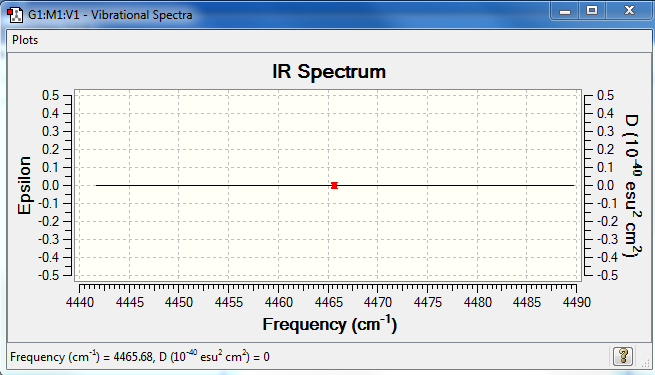
But as H2 molecule is a simple diatomic, consisting of 2 same atoms there is no change in dipole upon its vibration therefore no peaks are expected in the IR spectrum.
Predicted IR spectrum of H2
Charges
Charge on the H atoms - 0.000
N2 molecule
Summary
•molecule name: Dinitrogen/Molecular Nitrogen
•calculation method: B3LYP
•basis set: 6-31G(d,p)
•final energy E(RB3LYP) in atomic units (au): -109.52412868
•RMS gradientː 0.00000060
•the point group of your molecule: D∞h
Bond length (A) = 1.10550
Bond angle (Degrees)= 180 (Linear)
Item table
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES Predicted change in Energy=-3.401018D-13
Molecular model
Optimized Nitrogen molecule |
My N2 molecule link is here
Vibration modes
According to (3N - 5) rule only (6-5)= 1 mode of vibration expected, which is confirmed by the model computed. As the N atoms are heavier than H atoms, the frequency of vibration is lower for N2 molecule rather than H2.
But as N2 molecule is a simple diatomic, consisting of 2 same atoms there is no change in dipole upon its vibration therefore no peaks are expected in the IR spectrum.
Predicted IR spectrum of N2 molecule
Charges
Charge on the N atoms - 0.000
Energy calculations
Energies in atomic units
•E(NH3)= 56.55776873
•2*E(NH3)= 113.1155375
•E(N2)= 109.52412868
•E(H2)= 1.17853936
•3*E(H2)= 3.53561808
•ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= 0.05579074
Energies in kJ/mol
•ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= 146.4795978 = 146.48
This calculation was performed using Hess Cycle composed of standard enthalpies of formation of molecules involved in reaction.
The gaseous atoms (LHS) of the reaction are more stable asː The reaction is endothermic and the change in entropy is negative (as from 3 moles of gaseous molecules only 2 moles of gaseous molecules is made) therefore Gibbs energy is negative and reaction is not spontaneous; Thus the reaction will need an energy input in order to proceed.
Small molecule investigation - HCl
Summary
•molecule name: Hydrogen Chloride
•calculation method: B3LYP
•basis set: 6-31G(d,p)
•final energy E(RB3LYP) in atomic units (au): -460.80077875
•RMS gradientː 0.00005211
•the point group of your molecule: C∞v
Bond length (A) = 1.28599
Bond angle (Degrees) = 180 (Linear)
Item table
Item Value Threshold Converged? Maximum Force 0.000090 0.000450 YES RMS Force 0.000090 0.000300 YES Maximum Displacement 0.000139 0.001800 YES RMS Displacement 0.000197 0.001200 YES Predicted change in Energy=-1.256951D-08
Molecular model
Optimized Hydrogen Chloride molecule |
My HCl molecule link is here
Vibration modes
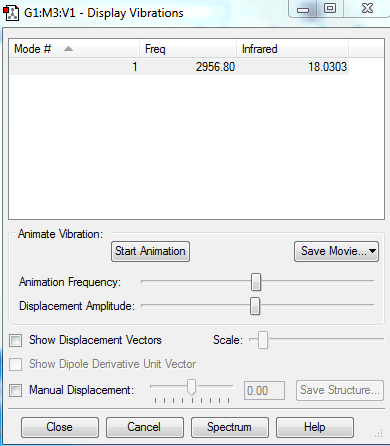
Expected number of modes of vibrations (3N-5)ː 6-5 = 1
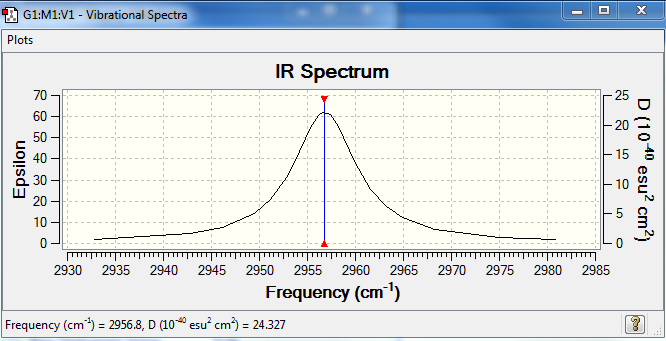
As HCl has a dipole, therefore a change in dipole is observed, which produces one peak in IR spectroscopy. The frequency is 2956.80, which indicates the high energy bond stretching.
Predicted IR spectrum of HCL
Charges
Charge on H atom = 0.284 Charge on Cl atom = -0.284
As expected, Cl atom is negatively charged and H atom is (equally) positively charged. According to Pauling table of electronegativity, Cl = 3.16 and H = 2.20. Cl is therefore more electronegative than H, attracting the shared electrons closer to its nucleus, which creates dipole and results in charge stated above.
Molecular Orbitals
In this diatomic molecule H's electron configuration [1s1] and Cl's [1s22s22p63s23p5]. H only atomic orbital (1s) has much higher energy than all of the Cl atomic orbitals. Cl atoms 1s, 2s, 2p and 3s orbitals are too deep in energy (largely stabilized) so that they can not mix with h 1s orbital (too big difference in energies). Thus the only atomic orbitals which are at appropriately close energy levels for mixing are Cl 3p orbitals and H 1s orbital.
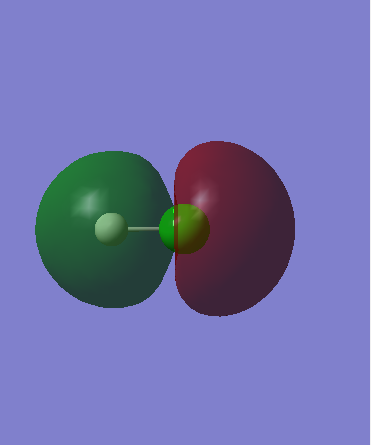
Only one of the 3 occupied 3p orbitals of Cl atom has appropriate geometry for constructive/disruptive interference with H 1s (ie is in phase), which assumed to be 3pz orbital. Orbital 7 diagram therefore represents the constructive overlap between 1s and 3pz atomic orbitals to produce a sigma molecular orbital. It has lower energy than both 1s and 3pz atomic orbitals therefore is stabilized. A shared pair of electrons is situated in this orbital.
As Cl atom is more electronegative than H atom, the sigma orbital will lie closer to Cl atom therefore will have greater 3pz character, which is seen from the Molecular orbital representation. It means that the shared electrons from the covalent bond will be primarily situated on Cl atom creating the dipole.
Orbital 7 - bonding sigmaː Energy = -0.47433
At the next energy level there are 2 filled non-bonding orbitals - 3px and 3py Cl atomic orbitals. They have 2 spin paired electrons each and are degenerate ie have the same energy. As they are the last filled orbitals at the highest energy level, either of them can act as a HOMO in reactions.
Orbital 8 (HOMO) - 3pxː Energy = -0.33163
Orbital 9 (HOMO) - 3pyː Energy = -0.33163
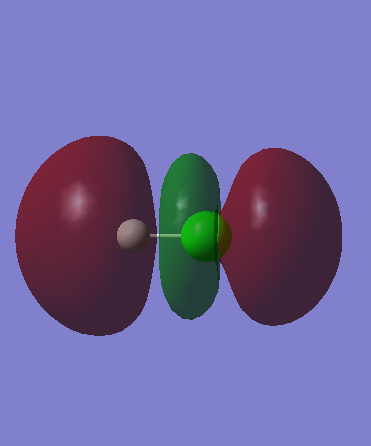
The LUMO of this molecule is an anti-bonding sigma orbital from 1s and 3pz overlap, which is represented by Orbital 10 diagram. It has higher energy than the previous orbitals and is closer to H atom therefore resembles 1s orbital character to the greater extent than 3pz. As it is produced from destructive interference there are 2 nodes present reinforcing the idea of the highest molecular orbital energy.
Orbital 10 (LUMO) - antibonding sigmaː Energy = 0.01366
For the size comparison, 2s (Orbital 2) and 3s (Orbital 6) Cl atom atomic orbitals are shown below. Their high energies show the impossibility in overlap with 1s H atom orbital. 2s orbital is at lower energy than 3s orbital hence is situated closer to the nucleus. Its size is much smaller as well for the same reason, due to greater coulombic attraction from nucleus as lower shielding from electrons from previous orbitals.
Orbital 2ː Energy = -9.47437
Orbital 6ː Energy = -0.84773
The possible explanation for the distorted shape of HCl 3s orbital might be due to Hydrogen 1s orbital being relatively close to it in Energy level to slightly contribute to its shape, but still too far for proper overlap. Another explanation could be the fact that it is still only a theoretically computed experimental module, which does not completely express the real nature of orbitals.
Small molecule investigation - CH3OH
Summary
•molecule name: Methanol
•calculation method: B3LYP
•basis set: 6-31G(d,p)
•final energy E(RB3LYP) in atomic units (au): -115.72136423
•RMS gradientː 0.01151827
•the point group of your molecule: Cs
Bond length (A)ː C-H = 1.07000; C-O = 1.43000, O-H = 0.96000
Bond angle (Degrees)ː H-C-H = 109.471; H-C-O = 109.471; C-O-H = 109.500
Item table
Item Value Threshold Converged? Maximum Force 0.000038 0.000450 YES RMS Force 0.000020 0.000300 YES Maximum Displacement 0.000492 0.001800 YES RMS Displacement 0.000239 0.001200 YES Predicted change in Energy=-1.606794D-08
Molecular model
Optimized Methanol molecule |
My Methanol molecule link is here
Vibration modes
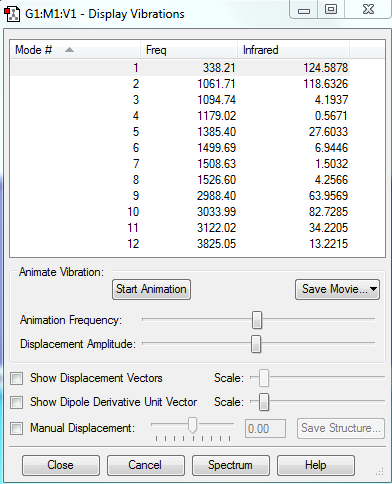
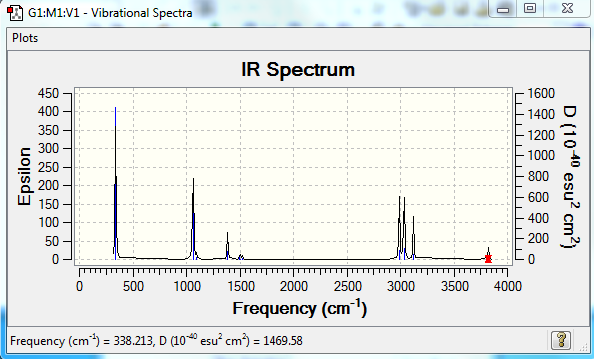
Expected number of modes of vibrations (3N-6) - 6 = 12
Modes 1-8 are bending modes of vibration; 9-12 are stretching modes of vibration. The larges frequency of vibration belongs to stretching of O-H bond as O is the heaviest atom with the strongest -H bond which therefore requires the most energy for vibration.
The predicted IR spectrum is shown below. Not all of the 12 peaks apper on the diagram asː1. Not all of the vibrations produce a change in dipole (eg stretch 9) 2. Some of the modes have very similar energies/frequencies thus it is harder to distinguish them on the spectrum.