Rep:Mod:KA671Q
Part 1: Learning how to carry out calculations
A note on accuracy
All energy values stated have an error of ~10kJ/mol, which equates to ~0.0038a.u. All discussion regarding energy values will use values rounded to 3dp.
Vibrational frequencies are fairly inaccurate and should be treated with caution. This is because Gaussview uses a harmonic approximation for all vibrations, and will apply this to an anharmonic case. Frequencies will be quoted to the nearest integer. Vibrational intensities will also be supplied to the nearest integer.
Bond distances are accurate to ~0.01A, and so bond lengths will be given to 2dp
Bond angles are accurate to ~0.10, angles will be given to 1dp.
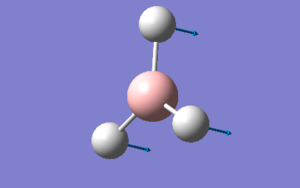
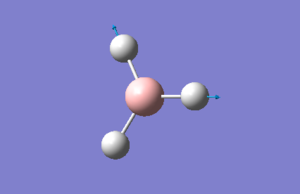
The humble BH3 molecule
BH3 was the first molecule which was created & subsequently optimised using GaussView (version 5.09). The .log output file can be found here: Media:OPTIMISEDBH3.LOG
The optimisation process involves finding the optimum position of the various atoms which make up a molecule for a given electronic structure. Most of the calculations used are based on Density Functional Theory (DFT). The optimisation calculation is carried out as follows. The B-O approximation is used, which means the atoms are assumed to be fixed in space. Then, the Schrodinger equation is solved for the electrons present in the molecule. Force = the first derivative of the potential energy, and it is this force (at a given potential energy) which is 'applied' to the molecule in order to reach a new structure (and thus a new potential energy). This process is repeated over and over until a minima in the potential energy of the molecule is reached, and therefore the next force applied = 0. The final positions of the atoms is the optimised structure.
The following table summarises the method used, and also the results of the process.
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 3-21G |
| Final energy (au) | -26.46226338 |
| Gradient (au) | 0.00020662 |
| Dipole moment | 0 |
| Point group | D3H |
| Calculation time | 2m 19s (laptop) |
Below is a copy of the 'item' table of the .log file.
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001643 0.001800 YES
RMS Displacement 0.001076 0.001200 YES
Predicted change in Energy=-1.018634D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
There are no surprises here. The point group, dipole & gradient are all what one would expect from a successful calculation and the item table shows that convergence was achieved, implying that F=-V/r. The optimum bond length for all BH bonds is 1.19 A.
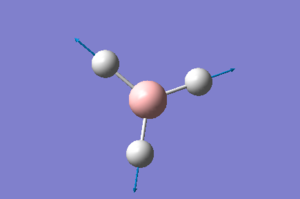
Using a higher level basis set to optimise BH3
The optimised BH3 molecule was then run through a second optimisation process, this time using the 6-31G(d,p) basis set. The .log output file can be seen here: media:OPTBH3_631G_DP.LOG
The basis sets seek to model atomic orbitals mathematically. The 6-31G basis set is more complex than the 3-21G basis set used in the first optimisation, and therefore gives a more accurate answer. However, the increased complexity means the calculation will take longer.
The results are summarised in the following tables.
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy (au) | -26.61532363 |
| Gradient (au) | 0.00000235 |
| Dipole moment | 0 |
| Point group | D3H |
| Calculation time | 4m 30s (laptop) |
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.305025D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
The results are as expected, with an acceptable gradient & convergence achieved. The process had to be run twice as the first calculation did not converge. The reason for this is unknown, but probably not something to be concerned about going forwards.
Optimised bond distance & angle were found using the 'inquiry' tool:
Bond length (B-H): 1.19 A
Bond angle: 1200
Note: the estimated bond length using a more complicated basis set (details contained in citation)[1] is 1.18 A, slightly different from my value. This is evidence of the limitations associated with the methods used.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedref1
Introducing psuedo-potentials & larger basis sets - TiBr3
This calculation was the first which was run on the HPC service. The .log output can be found here: [1]
Pseudopotentials are designed to try to mathematically model the core electrons of an atom - all non-valence electrons can be represented by a single pseudopotential approximation, which allows us to focus on the more important bonding electrons. Trying to calculate what effect the interaction of every single electron in a species such as TlBr3 has on its structure would be an impossible task given our current computing capabilities, so gross approximations such as this are vital.
As before, all relevant info is summarised in the following tables:
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | LANL2DZ |
| Final energy (au) | -91.21807632 |
| Gradient (au) | 0.00082814 |
| Dipole moment | 0 |
| Point group | D3H |
| Calculation time | 17 seconds (HPC) |
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-1.541780D-14
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Bond length (Ti-Br): 2.66 A
Bond angle: 1200
A bond length for Ti-Br could not be found (after all, Ti is not often worked with due to its toxic nature), but an experimental value for the bond length of Hg-Br was found to be 2.62 A. Given that this value is close to my value of 2.66 A, and given that Hg is next to Ti in the periodic table, this seems like good evidence that my value is reasonable.[1]
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedref2
A mixture of basis-sets and psuedo-potentials - BBr3
This optimisation process was run on the HPC. The completed calculation files can be found here: [2]
As before, relevant data is tabulated below.
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | Gen |
| Final energy (au) | -64.42903745 |
| Gradient (au) | 0.00000382 |
| Dipole moment | 0 |
| Point group | D3H |
| Calculation time | 18 seconds (HPC) |
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027252D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Bond length: 1.93 A
Bond angle: 1200
A literature value for BBr3 was found to be 1.90 A[1], evidence that my value is probably correct but also more evidence of the limitations of the methods used.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedref3
Analysis of results
| Molecule (XY3) | Bond length (XY) (A) |
|---|---|
| BH3 | 1.19 |
| TlBr3 | 2.67 |
| BBr3 | 1.93 |
First, it's useful to discuss what a bond is. The bond length (or, more accurately, mean bond length) between the two atoms in a diatomic is simply the distance between the two atoms at which the energy of the system happens to be at its lowest point. There are several factors which influence the bond length:
- The number of electrons involved in the bond (bond order)
- The size of the constituent atoms
- The type of orbitals involved in the bond (1s will overlap very well with 1s, leading to a short bond. It will overlap less well with 2s, leading to a somewhat longer bond. It will overlap even less well with 2p, leading to an even longer bond, etc.)
- The relative electronegativities of the constituent atoms (and therefore the type of bond)
And so on. A strong bond involving atoms of roughly the same size will be shorter than a weak bond.
BH3 has strong, short BH bonds because of two factors. Both B and H are relatively small atoms, and the orbitals involved in the bonding (2s on boron, 1s on hydrogen) overlap relatively well. Boron (2.0) and hydrogen (2.1) also have very similar electronegativities, which strengthens the bond between them (and therefore shortens it relative to bonds between constituents with large differences in electronegativity).
The BBr3 bonds are much larger than the bonds in BH3. This is because Br has a much larger atomic radius compared with H, but also because the orbitals involved in the bond have poor overlap (2s of boron, 4p/3d of Br). The two atoms also have quite different electronegativities (B = 2.0, Br = 2.8), which weakens (and lengthens) the bond somewhat.
While Boron and Thallium are in the same group, they are hardly alike. Tl's atomic radii is more than double that of boron, and its valence electrons are 6p rather than boron's 2p. The fact that the B-Br bond is much smaller compared with the Tl-Br bond is no surprise given the enormous size difference.
Frequency analysis - BH3
Link to the completed frequency analysis of BH3: media:MAX_BH3_FREQ.LOG
Frequency analysis is carried out to make sure the optimisation was a success. The optimisation process can fail, for example, if it finds a local minima in potential energy rather than the overall minima. If that happens, the frequency analysis will show it. Some frequencies will be negative if the optimisation process has not been successful.
The 'low frequencies', which will be included in this wiki for every frequency analysis done, represent the motion of the centre of mass.
The low frequencies for this calculation: (within +/- 15cm-1)!
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824
Low frequencies --- 1163.0003 1213.1853 1213.1880
Tabulating the various vibrational modes:
| Vibrational mode | Frequency (cm-1) | Infrared |
|---|---|---|
| 1 | 1163 | 93 |
| 2 | 1213 | 14 |
| 3 | 1213 | 14 |
| 4 | 2582 | 0 |
| 5 | 2715 | 126 |
| 6 | 2715 | 126 |
Images & description of vibrational motion:
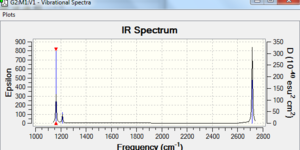
The IR spectrum of the optimised molecule is shown to the right. There appears to be 3 peaks, which does not make intuitive sense given that the molecule has 6 vibrational modes. In fact, modes 2&3 and modes 5&6 have the same vibrational frequency, and so are indistinguishable on the IR spectra. All 4 peaks associated with those vibrations are in fact present, as is the peak associated with vibrational mode 1.
Mode 4, which has a vibrational frequency of 2582 cm-1 is not present, this is because that particular frequency for the optimised BH3 has an associated infrared intensity of 0.
Frequency analysis of TlBr3 & comparison with BH3
Link to the completed frequency analysis of TlBr3 (carried out on HPC): [3]
Relevant data presented below. Frequencies in cm-1.
Low frequencies --- -3.4046 -0.0026 -0.0004 0.0016 3.9514 3.9514
Low frequencies --- 46.4301 46.4304 52.1468
| Vibrational mode | Frequency(cm-1) | Infrared |
|---|---|---|
| 1 | 46 | 4 |
| 2 | 46 | 4 |
| 3 | 52 | 6 |
| 4 | 165 | 0 |
| 5 | 210 | 25 |
| 6 | 210 | 25 |
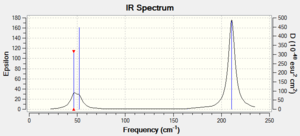
Comparison of BH3 & TlBr3 vibrational frequencies. Frequencies in cm-1.
| Vibrational mode | BH3 vibrations | TlBr3 vibrations |
|---|---|---|
| 1 | 1163 | 46 |
| 2 | 1213 | 46 |
| 3 | 1213 | 52 |
| 4 | 2582 | 165 |
| 5 | 2715 | 211 |
| 6 | 2715 | 211 |
The obvious difference between the two molecules' vibrational frequencies is that the heavier TlBr3 has much lower frequencies compared with BH3. This is to be expected as TlBr units have a much greater mass compared with BH units, therefore the vibrations are much slower (although I note that this difference is not represented in Gaussview).
The IR spectra comparison is interesting. On the one hand, they are similar in that the frequency ratio between the three peaks is approximately the same on both spectra. This can be demonstrated by dividing the gap between the second two peaks by the gap between the first two peaks. Note that the distance between the peaks have been roughly estimate, an accurate answer is not necessary to prove the point.
BH3: 1500/60 = 19.2
TlBr3: 150/8 = 18.8
The peaks do however differ in their shape. While the BH3 peaks are sharp and well defined, the TlBr3 peaks are wider and more diffuse. This is partly due to the difference in scale, but also because of the larger nature of TlBr, which leads to more spread out peaks.
The perfectly symmetrical stretching mode, #4, has a peak magnitude of 0 for both molecules. This is because the overall dipole of the vibration is 0, so will not be observed on an IR spectra. The stretches associated with both molecules are at a higher frequency than the bending vibrations, which is expected as E(streches)>E(bends).
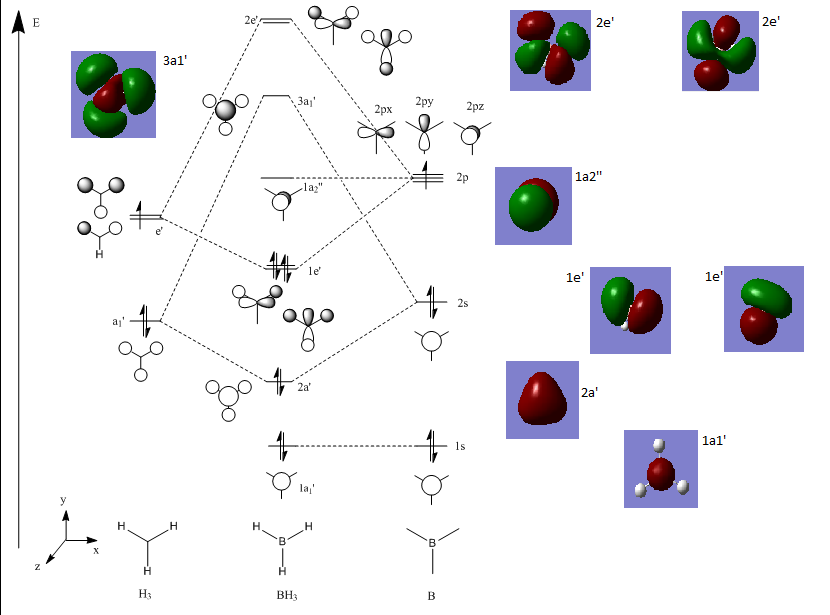
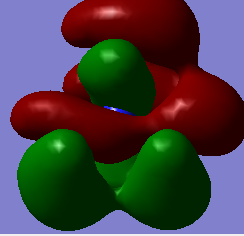
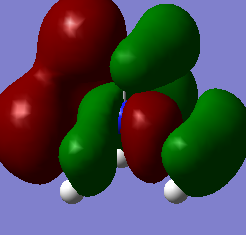
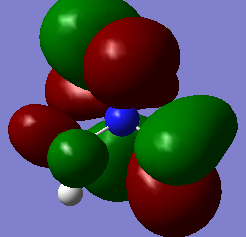
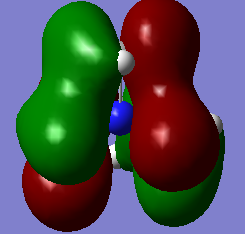
Molecular orbitals of BH3
Below is an MO diagram of BH3 made in chemdraw, along with the 'real' MOs computed by Gaussview, and a link to the completed population analysis stored on D-space
Population analysis[4]
(Based on 2nd year MO lecture notes)
The LCAOs are roughly identical to the computed MOs, which shows the usefulness of qualitative MO theory in describing MOs.
NH3 - NBO analysis
.Log output of the optimisation calculation can be found here: MEDIA:NH3OPT.LOG, and a link to the vibrational analysis output log here: MEDIA:NH3VIB.LOG
Note: I was having trouble logging into the HPC service while attempting these calculations, so they were done on the laptop instead.
Item Value Threshold Converged?
Maximum Force 0.000095 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000798 0.001800 YES
RMS Displacement 0.000466 0.001200 YES
Predicted change in Energy=-1.250846D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.0181 -DE/DX = -0.0001 !
! A1 A(2,1,3) 105.6756 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.6917 -DE/DX = 0.0001 !
! A3 A(3,1,4) 105.6917 -DE/DX = 0.0001 !
! D1 D(2,1,4,3) 111.7426 -DE/DX = 0.0001 !
--------------------------------------------------------------------------------
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy (au) | -56.55776837 |
| Gradient (au) | 0.00006944 |
| Dipole moment | 1.8492 |
| Point group | CS |
| Calculation time | 4 seconds (Laptop) |
N-H bond length: 1.02A
H-N-H bond angle: 106.00
The point group has come out as CS, whereas is should be C3v. An effort was made to restrict the point group as with TlBr3 optimisation, but this was unsuccessful as gaussview did not provide an option to restrict the point group to C3v
The frequency analysis was as expected (all positive), which confirms that the optimisation process went according to plan. The vibrations were roughly analogous with the planar BH3 molecule discussed earlier, except that vibration 4 showed up on the spectrum because the overall dipole of the vibration was >0 (due to the lone pair causing the molecule to adopt a non-planar configuration).
The low frequencies associated with this molecule are shown below.
Low frequencies --- -13.7087 -6.1355 -0.0010 -0.0008 0.0008 21.0007 Low frequencies --- 1092.3352 1694.3311 1694.4269
The .chk output file from the population analysis can be found here: MEDIA:NH3pop.chk, the .log can be found here: MEDIA:NH3POP.LOG
Picture of the charge on NH3 in thumbnail. Colour range used: -1 to +1.
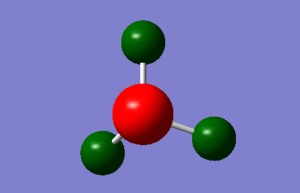
The specific NBO charges for the atoms are as follows:
Each hydrogen atom has a charge of +0.375
The Nitrogen atom has a charge of -1.125
Association energies - NH3BH3
First, the molecule was optimised and then a frequency analysis was run. Log output for the optimisation can be found here: MEDIA:NH3BH3_OPT.LOG. Log output for the frequency analysis:
The relevant details can be found below,
Item Value Threshold Converged?
Maximum Force 0.000122 0.000450 YES
RMS Force 0.000058 0.000300 YES
Maximum Displacement 0.000557 0.001800 YES
RMS Displacement 0.000308 0.001200 YES
Predicted change in Energy=-1.704689D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.2101 -DE/DX = -0.0001 !
! R2 R(2,7) 1.21 -DE/DX = -0.0001 !
! R3 R(3,7) 1.21 -DE/DX = -0.0001 !
! R4 R(4,8) 1.0186 -DE/DX = -0.0001 !
! R5 R(5,8) 1.0186 -DE/DX = -0.0001 !
! R6 R(6,8) 1.0186 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6681 -DE/DX = -0.0001 !
! A1 A(1,7,2) 113.8759 -DE/DX = 0.0 !
! A2 A(1,7,3) 113.8731 -DE/DX = 0.0 !
! A3 A(1,7,8) 104.5959 -DE/DX = 0.0 !
! A4 A(2,7,3) 113.8787 -DE/DX = 0.0 !
! A5 A(2,7,8) 104.5963 -DE/DX = 0.0 !
! A6 A(3,7,8) 104.5924 -DE/DX = 0.0 !
! A7 A(4,8,5) 107.87 -DE/DX = 0.0 !
! A8 A(4,8,6) 107.8697 -DE/DX = 0.0 !
! A9 A(4,8,7) 111.0282 -DE/DX = 0.0 !
! A10 A(5,8,6) 107.8736 -DE/DX = 0.0 !
! A11 A(5,8,7) 111.0262 -DE/DX = 0.0 !
! A12 A(6,8,7) 111.0281 -DE/DX = 0.0 !
! D1 D(1,7,8,4) 179.9802 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -60.0216 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 59.9811 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -60.0187 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 59.9795 -DE/DX = 0.0 !
! D6 D(2,7,8,6) 179.9823 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 59.984 -DE/DX = 0.0 !
! D8 D(3,7,8,5) 179.9822 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -60.015 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
| Calculation type | FOPT |
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy (au) | -83.22468988 |
| Gradient (au) | 0.00005844 |
| Dipole moment | 5.5649 |
| Point group | C1 |
| Calculation time | 1m 49s (laptop) |
Vibrational analysis showed that all vibrations of the optimised molecule are positive, this is evidence that the optimisation was successful.

Energy (in AU) of the relevant molecules: (note - all energy values compared here have been obtained using identical basis sets & computational methods)
NH3 = -56.558
BH3 = -26.615
NH3BH3 = -83.225
The change in energy when going from 1 molecule of BH3 and 1 of NH3 to a molecule of NH3BH3 = -83.225 - (-56.558 - 26.615) = -0.052
1 AU = 2625.50 kJ/mol
-0.052*2625.5 = -136 kJ/mol. The magnitude of this value represents the bond dissociation energy of NH3BH3
According to a US dept. of energy study[1], the true bond dissociation energy of the molecule is 109kJ/mol, which is a roughly ballpark figure. A more recent (2009) journal entry[2] has an answer of 130 kJ/mol, much closer to the answer found by calculation.
The figure given by the calculation is clearly an answer to the question of the bond dissociation energy of NH3BH3, although parhaps not all that accurate an answer.
Part 2 - Ionic liquids & designer solvents
[N(CH3)4]+
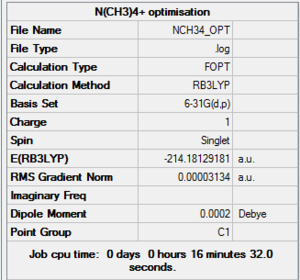
NOTE: There were problems accessing the HPC website (http://scanweb.cc.imperial.ac.uk/uportal2/index.php), so all calculations were run on the laptop rather than on the HPC.
Optimisation & frequency analysis log outputs for [N(CH3)4]+MEDIA:NCH34_OPT.LOG, MEDIA:NCH34_FREQ.LOG
Item table & low frequencies below, summary of optimisation to the right:
Item Value Threshold Converged? Maximum Force 0.000090 0.000450 YES RMS Force 0.000026 0.000300 YES Maximum Displacement 0.000908 0.001800 YES RMS Displacement 0.000241 0.001200 YES Predicted change in Energy=-9.969287D-08 Optimization completed. -- Stationary point found.
Low frequencies --- -0.0011 -0.0010 -0.0009 14.0798 19.0341 24.8016 Low frequencies --- 193.8515 289.8842 295.0678
[P(CH3)4]+
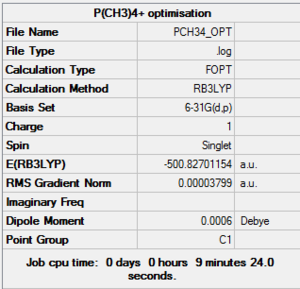
Optimisation & frequency analysis log outputs for [N(CH3)4]+MEDIA:PCH34_OPT.LOG, MEDIA:PCH34_FREQ.LOG
Item table & low frequencies below, summary of optimisation to the right:
Item Value Threshold Converged? Maximum Force 0.000078 0.000450 YES RMS Force 0.000025 0.000300 YES Maximum Displacement 0.000951 0.001800 YES RMS Displacement 0.000289 0.001200 YES Predicted change in Energy=-1.292866D-07 Optimization completed. -- Stationary point found.
Low frequencies --- -15.5125 -1.8148 -0.0015 -0.0003 0.0031 11.8077 Low frequencies --- 154.5280 187.3969 191.2187
[S(CH3)3]+
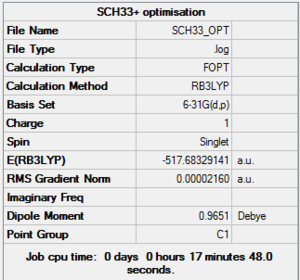
Optimisation & frequency analysis log outputs for [S(CH3)3]+MEDIA:SCH33_OPT.LOG, MEDIA:SCH33_FREQ.LOG
Item table & low frequencies below, summary of optimisation to the right:
Item Value Threshold Converged? Maximum Force 0.000067 0.000450 YES RMS Force 0.000021 0.000300 YES Maximum Displacement 0.000632 0.001800 YES RMS Displacement 0.000245 0.001200 YES Predicted change in Energy=-1.355854D-07 Optimization completed. -- Stationary point found.
Low frequencies --- -17.5556 -0.0036 -0.0027 0.0016 14.2254 36.4170 Low frequencies --- 164.5978 194.6214 208.4818
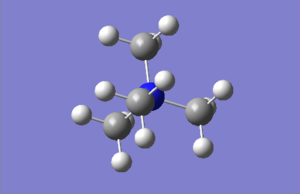
Structure comparison (click to enlarge)
| [N(CH3)4]+ | [P(CH3)4]+ | [S(CH3)3]+ |
|---|---|---|
 |
 |
 |
Both of the first two structures adopt simple tetrahedral shapes, while the sulfur-containing molecule adopts a very different configuration.
The two tetrahedral molecules have an identical C-X-C bond angle of 109.50, which one would expect. There is of course a difference in the C-X bond angle of the two molecules, the N-C bond being 1.51A in length while the C-P bond is 1.82A. This is due to the larger atomic radius of phosphorus when compared with nitrogen. In terms of bond strength, theory states that the N-C bond should be stronger as the nitrogen bonding orbitals overlap very well with the carbon ones, whereas phosphorus-carbon bonds are 3p-2p, and therefore not as strong. The shorter bond length of C-N compared with C-P bears this out.
The sulfur-containing molecule involves a lone pair situated on the sulfur atom, which, in accordance with VSEPR theory, has a larger relative repulsive effect on bonding pairs. This leads to a distorted trigonal pyramidal configuration. The central C-S-C bond angle, 102.70, is smaller than the traditional example of a trigonal pyrimidal structure, NH3, which has a bond angle of 107.80. This is why the structure is known as distorted. The effect is due to the lower electron density of P compared with N. The bonding pair-bonding pair repulsion in NH3 > [P(CH3)3]+ because a relatively greater proportion of the electrons in the N-H bonded pairs are situated at the N (greater electronegativity), compared to the P-C bonds in [P(CH3)3]+. This is enough to overhaul the greater steric repulsive effect of CH3 groups compared with H groups, leading to the distorted trigonal pyrimidal structure.
Population analysis
The three .log & .chk outputs for the population analyses can be found below
[N(CH3)4]+: media:NCH34_NBO.LOG, media:NCH34_nbo.chk
[P(CH3)4]+: media:PCH34_NBO.LOG, media:PCH34_nbo.chk
[S(CH3)3]+: media:SCH33_NBO.LOG, media:SCH33_nbo.chk
| [N(CH3)4]+ | [P(CH3)4]+ | [S(CH3)3]+ |
|---|---|---|
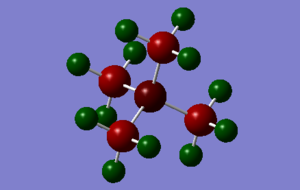 |
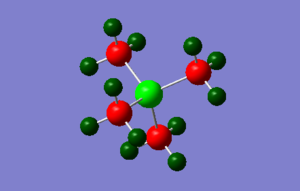 |
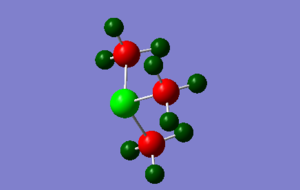 |
All NBO analysis was done under the following conditions: type: NBO, colour range, -1 to 1. Specific charges on each atom are described below. The sulfur containing molecule is more complex than the other two and is accompanied by a picture.

N(CH3)4]+: H (12): +0.269, C (4): -0.483, N (1): -0.295
P(CH3)4]+: H (12): +0.298, C (4): -1.060, P (1), +1.667
[S(CH3)3]+: Type 1 H (6): +0.297, Type 2 H (3): +0.279, C (3): -0.845, S (1): +0.917
As expected, the sum of all charges for each molecule totals +1.
The most useful comparison to be made is of the three central atoms, as they are all in roughly similar environments. One would expect that the amount of positive charge on the central atom to be negatively correlated with the electronegativity of the atom in question (as electronegativity rises, charge decreases), this is borne out by the data. The most electronegative atom of the three, N, has the most negative charge, the second most, S, the second most negative charge and P, the most electropositive, has the least negative charge. This electronegativity analysis falls down when the whole molecule is considered however, and this is most easily demonstrated by looking at the nitrogen-containing molecule.
The Pauling electronegativity values of the three atoms which make up [N(CH3)4]+ are as follows: H = 2.1, C = 2.55, N = 3.0. If the electronegativity argument was enough to explain everything regarding charge on these molecules, then carbon would bear less negative charge compared to N, because the electronegativity of N > C. The opposite is in fact the case, the charge on N is -0.295 while the charge on C is -0.483.

In order to investigate this, a molecule of HCN was drawn up in Gaussview, optimised, a frequency analysis was done to confirm the success of the optimisation process, and NBO analysis was done to see where the charge was distributed. .logs of the optimisation, frequency analysis & pop calculation: MEDIA:HCN_OPT.LOG, MEDIA:HCN_FREQ.LOG, MEDIA:HCN_POP.LOG - low freqencies and item list are displayed further down. the results are shown to the right, and are in line with the electronegativity prediction. Most of the electron density in the CN bond is situated on the more electronegative N atom.
The only meaningful difference between this idealised CN bond and the CN bonds in N(CH3)4]+ is the fact that the latter molecule has an overall positive charge. The answer to the original question of why the carbon atoms bear more negative charge than the N is explained by this. The + charge means that the central nitrogen 'feels' the positive charge more than the other atoms, which has the effect of making it more electropositive than the Pauling table would indicate.
A similar phenomenon can be seen on the phosphorus-containing molecule. The electronegativity of P (2.19) > H (2.10), but the P atom has a much greater positive charge compared with the H atoms. This is again because of the overall positive charge on the molecule and the effect it has on the centrally located the P atom.
The carbon atoms follow a simple and predictable pattern; the more electronegative the central atom, the less negative charge there is on the carbon atoms.
The hydrogens also follow a simple pattern. The more electron density is on the carbons, the more positive the hydrogens are. To relate this to the central atom, a more electronegative central atom will result in a less positively charged set of hydrogens. This is an effect that is widely exploited in organic chemistry, for example the generation of enolates by removal of an alpha proton.
The hydrogens on the sulfur-containing molecule bear closer inspection, since it is the only molecule of the three which contains more than 1 type of proton. There is no doubt that this is due to the lone pair on the sulfur. If the LP is considered to be a bond, then a sterioelectronic argument can be made to rationalise the data. The less positively charged 'type 2' proton-carbon bonds are 1800 to the hypothetical 'lone pair bond', which means that electron density can be donated into the cis-C-H bond from the lone pair, not something which can occur with the non-cis H-C bonds. The greater electron density decreases the negative charge on the hydrogen. Another effect of this would be to strengthen the cis CH bonds, and therefore shorten the length of those bonds, and this is indeed the case. The 'type 2' C-H bonds are all between 1.09139-1.09140A in length, while the non-cis CH bonds are all between 1.09158-1.09160A. NOTE: These values are beyond the tolerance of the errors associated with bond lengths, so the bond length evidence must be treated with caution.
Item table & low frequencies for HCN:
Item Value Threshold Converged? Maximum Force 0.000079 0.000450 YES RMS Force 0.000039 0.000300 YES Maximum Displacement 0.000128 0.001800 YES RMS Displacement 0.000078 0.001200 YES Predicted change in Energy=-7.431214D-09 Optimization completed. -- Stationary point found.
Low frequencies --- -0.0004 -0.0003 0.0016 5.3964 5.3964 900.9087 Low frequencies --- 900.9087 2175.2002 3496.2334
Quarternary ammonium cations are usually represented with the positive charge resting on the N. This makes sense as a relative charge indicator (relative to the usual charge on N), but does not give an accurate picture of where the positive charge is actually located in the molecule. What the charge really represents when it is placed on the nitrogen: 'in this particular arrangement, this atom has less electron density situated on it than you might ordinarily expect this type of atom to have'. In fact, the positive charge is distributed evenly among the H atoms of the molecule, but it would be impractical to draw this out.
NBO population analysis
A population analysis was run on each molecule and the relevant results are displayed below.
[N(CH3)4]+
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99119) BD ( 1) C 1 - H 2
( 63.47%) 0.7967* C 1 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 0.1701
0.0166 0.5718 -0.0201 -0.6154 -0.0043
0.0040 -0.0031 -0.0194 -0.0100 0.0044
( 36.53%) 0.6044* H 2 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0075 -0.0125 0.0178
2. (1.99118) BD ( 1) C 1 - H 3
( 63.47%) 0.7967* C 1 s( 26.41%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5139 0.0032 -0.0004 0.6835
0.0075 -0.1260 -0.0077 0.5016 -0.0241
-0.0031 0.0196 -0.0032 0.0105 0.0033
( 36.53%) 0.6044* H 3 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0203 0.0048 -0.0098
3. (1.99118) BD ( 1) C 1 - H 4
( 63.47%) 0.7967* C 1 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 -0.5140 -0.0032 0.0004 0.2783
-0.0245 0.7388 -0.0031 0.3336 0.0093
-0.0137 -0.0059 -0.0112 0.0111 0.0073
( 36.53%) 0.6044* H 4 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0006 -0.0036 -0.0200 -0.0109
4. (1.98452) BD ( 1) C 1 - N 17
( 33.65%) 0.5801* C 1 s( 20.78%)p 3.81( 79.06%)d 0.01( 0.16%)
0.0003 0.4552 -0.0237 0.0026 -0.6506
-0.0276 0.3314 0.0141 0.5061 0.0215
-0.0192 -0.0293 0.0149 0.0140 -0.0005
( 66.35%) 0.8146* N 17 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 0.5000 -0.0007 0.0000 0.6340
-0.0001 -0.3229 0.0001 -0.4934 0.0001
-0.0084 -0.0129 0.0066 0.0061 -0.0002
5. (1.99118) BD ( 1) C 5 - H 6
( 63.47%) 0.7967* C 5 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 0.1812
0.0048 0.5386 0.0136 -0.6416 0.0221
0.0039 -0.0061 -0.0181 -0.0051 0.0109
( 36.53%) 0.6044* H 6 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0059 -0.0173 0.0141
6. (1.99118) BD ( 1) C 5 - H 7
( 63.47%) 0.7967* C 5 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 -0.6418
0.0193 0.2997 0.0178 0.4826 0.0022
-0.0088 -0.0169 0.0054 0.0113 -0.0013
( 36.53%) 0.6044* H 7 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 0.0145 -0.0114 -0.0138
7. (1.99119) BD ( 1) C 5 - H 8
( 63.47%) 0.7967* C 5 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 0.6953
-0.0043 -0.1587 0.0259 0.4753 0.0023
-0.0089 0.0166 -0.0060 0.0115 -0.0017
( 36.53%) 0.6044* H 8 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0186 0.0000 -0.0136
8. (1.98452) BD ( 1) C 5 - N 17
( 33.65%) 0.5801* C 5 s( 20.77%)p 3.81( 79.06%)d 0.01( 0.16%)
0.0003 0.4552 -0.0237 0.0026 -0.2654
-0.0113 -0.7687 -0.0326 -0.3576 -0.0152
0.0182 0.0085 0.0245 -0.0232 -0.0104
( 66.35%) 0.8146* N 17 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 0.5000 -0.0007 0.0000 0.2588
0.0000 0.7491 -0.0001 0.3487 0.0000
0.0080 0.0037 0.0108 -0.0102 -0.0046
9. (1.99118) BD ( 1) C 9 - H 10
( 63.47%) 0.7967* C 9 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 -0.5140 -0.0032 0.0004 0.6273
-0.0053 -0.3406 0.0234 -0.4744 -0.0111
0.0136 0.0140 -0.0096 -0.0063 0.0031
( 36.53%) 0.6044* H 10 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0006 -0.0165 0.0055 0.0151
10. (1.99118) BD ( 1) C 9 - H 11
( 63.47%) 0.7967* C 9 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 -0.2538
-0.0014 -0.7312 -0.0044 -0.3681 0.0260
0.0088 0.0058 0.0167 -0.0112 -0.0026
( 36.53%) 0.6044* H 11 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 0.0073 0.0211 0.0058
11. (1.99118) BD ( 1) C 9 - H 12
( 63.47%) 0.7967* C 9 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 0.7093
-0.0184 -0.1191 -0.0152 0.4661 0.0113
-0.0028 0.0168 -0.0010 0.0149 -0.0034
( 36.53%) 0.6044* H 12 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0166 0.0059 -0.0149
12. (1.98452) BD ( 1) C 9 - N 17
( 33.65%) 0.5801* C 9 s( 20.77%)p 3.81( 79.06%)d 0.01( 0.16%)
-0.0003 -0.4551 0.0237 -0.0026 -0.1943
-0.0082 -0.5765 -0.0245 0.6474 0.0275
-0.0100 0.0112 0.0333 0.0131 -0.0120
( 66.35%) 0.8146* N 17 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 -0.5000 0.0007 0.0000 0.1893
0.0000 0.5619 -0.0001 -0.6310 0.0001
-0.0044 0.0049 0.0146 0.0058 -0.0053
13. (1.99118) BD ( 1) C 13 - H 14
( 63.47%) 0.7967* C 13 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 -0.2373
-0.0175 -0.7532 0.0175 -0.3330 -0.0091
0.0075 0.0023 0.0124 -0.0160 -0.0073
( 36.53%) 0.6044* H 14 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 0.0096 0.0180 0.0108
14. (1.99118) BD ( 1) C 13 - H 15
( 63.47%) 0.7967* C 13 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 -0.6119
-0.0109 0.3192 -0.0015 0.5082 -0.0240
-0.0093 -0.0174 0.0096 0.0054 0.0036
( 36.53%) 0.6044* H 15 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 0.0189 -0.0086 -0.0100
15. (1.99119) BD ( 1) C 13 - H 16
( 63.47%) 0.7967* C 13 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 0.2110
-0.0255 0.5571 -0.0057 -0.6162 -0.0041
0.0086 -0.0093 -0.0174 -0.0065 0.0045
( 36.53%) 0.6044* H 16 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0015 -0.0145 0.0178
The data shows that ~66% of the electron density in the C-N bonds is provided by the N, while the C makes up the remaining 34%. This disparity is explained by the electronegativity argument, N is more electronegative so should in theory contribute more to the overall bond. The same argument explains the C-H disparity (63% C).
[P(CH3)3]+
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98386) BD ( 1) C 1 - H 2
( 64.78%) 0.8049* C 1 s( 24.88%)p 3.02( 75.07%)d 0.00( 0.04%)
-0.0001 0.4988 -0.0071 0.0005 0.3452
-0.0028 0.6567 -0.0059 0.4470 0.0210
0.0135 0.0054 0.0102 -0.0094 -0.0053
( 35.22%) 0.5934* H 2 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0080 -0.0151 -0.0140
2. (1.98385) BD ( 1) C 1 - H 3
( 64.78%) 0.8049* C 1 s( 24.88%)p 3.02( 75.07%)d 0.00( 0.04%)
0.0001 -0.4988 0.0070 -0.0005 0.8358
-0.0005 0.0840 0.0039 -0.2113 -0.0216
-0.0025 0.0025 0.0007 -0.0183 0.0090
( 35.22%) 0.5934* H 3 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 -0.0202 -0.0026 0.0083
3. (1.98385) BD ( 1) C 1 - H 4
( 64.78%) 0.8049* C 1 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0071 0.0005 0.4206
-0.0030 -0.7321 -0.0022 0.1934 0.0217
-0.0166 0.0005 -0.0018 -0.0083 -0.0091
( 35.22%) 0.5935* H 4 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0098 0.0181 -0.0079
4. (1.98030) BD ( 1) C 1 - P 17
( 59.57%) 0.7718* C 1 s( 25.23%)p 2.96( 74.68%)d 0.00( 0.08%)
0.0002 0.5020 0.0171 -0.0020 0.0696
-0.0013 0.1583 -0.0029 -0.8466 0.0155
0.0007 -0.0040 -0.0090 -0.0007 0.0273
( 40.43%) 0.6358* P 17 s( 25.00%)p 2.97( 74.15%)d 0.03( 0.85%)
0.0000 0.0001 0.5000 -0.0008 0.0000
0.0000 -0.0694 0.0001 0.0000 -0.1576
0.0002 0.0000 0.8437 -0.0012 0.0024
-0.0126 -0.0287 -0.0022 0.0868
5. (1.98387) BD ( 1) C 5 - H 6
( 64.79%) 0.8049* C 5 s( 24.89%)p 3.02( 75.07%)d 0.00( 0.04%)
-0.0001 0.4988 -0.0071 0.0005 0.3434
-0.0120 0.6588 0.0173 0.4453 -0.0063
0.0108 0.0114 0.0131 -0.0032 -0.0005
( 35.21%) 0.5934* H 6 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0065 -0.0186 -0.0099
6. (1.98385) BD ( 1) C 5 - H 7
( 64.78%) 0.8049* C 5 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
0.0001 -0.4987 0.0071 -0.0005 -0.0691
0.0112 -0.1593 -0.0187 0.8486 0.0028
0.0010 0.0063 0.0015 0.0001 -0.0197
( 35.22%) 0.5935* H 7 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 0.0000 0.0066 -0.0210
7. (1.98384) BD ( 1) C 5 - H 8
( 64.78%) 0.8049* C 5 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
0.0001 -0.4987 0.0070 -0.0005 0.8362
0.0088 0.0841 -0.0193 -0.2097 0.0056
-0.0080 0.0098 0.0035 -0.0136 0.0086
( 35.22%) 0.5934* H 8 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 -0.0216 0.0008 0.0043
8. (1.98030) BD ( 1) C 5 - P 17
( 59.57%) 0.7718* C 5 s( 25.23%)p 2.96( 74.68%)d 0.00( 0.08%)
-0.0002 -0.5020 -0.0171 0.0020 -0.4210
0.0077 0.7296 -0.0134 -0.1923 0.0035
0.0207 -0.0054 0.0094 0.0120 0.0124
( 40.43%) 0.6359* P 17 s( 25.00%)p 2.97( 74.15%)d 0.03( 0.85%)
0.0000 -0.0001 -0.5000 0.0008 0.0000
0.0000 0.4193 -0.0006 0.0000 -0.7273
0.0010 0.0000 0.1915 -0.0003 0.0658
-0.0173 0.0300 0.0381 0.0393
9. (1.98386) BD ( 1) C 9 - H 10
( 64.78%) 0.8049* C 9 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
0.0001 -0.4988 0.0071 -0.0005 -0.0692
0.0092 -0.1574 0.0176 0.8489 0.0094
-0.0029 0.0050 0.0106 0.0023 -0.0167
( 35.22%) 0.5934* H 10 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 0.0003 0.0012 -0.0220
10. (1.98385) BD ( 1) C 9 - H 11
( 64.78%) 0.8049* C 9 s( 24.88%)p 3.02( 75.07%)d 0.00( 0.04%)
-0.0001 0.4988 -0.0070 0.0005 0.4209
-0.0101 -0.7323 -0.0152 0.1921 -0.0122
-0.0152 0.0079 -0.0080 -0.0042 -0.0073
( 35.22%) 0.5934* H 11 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0087 0.0200 -0.0029
11. (1.98385) BD ( 1) C 9 - H 12
( 64.78%) 0.8049* C 9 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
0.0001 -0.4987 0.0070 -0.0005 0.8361
0.0067 0.0854 0.0170 -0.2095 0.0122
0.0008 0.0118 -0.0017 -0.0156 0.0068
( 35.22%) 0.5934* H 12 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 -0.0213 -0.0046 0.0033
12. (1.98030) BD ( 1) C 9 - P 17
( 59.57%) 0.7718* C 9 s( 25.24%)p 2.96( 74.68%)d 0.00( 0.08%)
0.0002 0.5021 0.0171 -0.0020 0.3438
-0.0063 0.6562 -0.0120 0.4446 -0.0082
0.0152 0.0103 0.0197 -0.0105 -0.0030
( 40.43%) 0.6358* P 17 s( 25.00%)p 2.97( 74.15%)d 0.03( 0.85%)
0.0000 0.0001 0.5000 -0.0008 0.0000
0.0000 -0.3425 0.0005 0.0000 -0.6540
0.0009 0.0000 -0.4432 0.0006 0.0483
0.0327 0.0625 -0.0335 -0.0095
13. (1.98385) BD ( 1) C 13 - H 14
( 64.79%) 0.8049* C 13 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4988 -0.0070 0.0005 0.4229
0.0207 -0.7310 0.0042 0.1925 -0.0060
-0.0105 0.0027 -0.0095 -0.0120 -0.0089
( 35.21%) 0.5934* H 14 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0133 0.0171 -0.0038
14. (1.98385) BD ( 1) C 13 - H 15
( 64.78%) 0.8049* C 13 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
0.0001 -0.4988 0.0070 -0.0005 -0.0710
-0.0217 -0.1585 -0.0018 0.8486 0.0032
0.0004 -0.0032 0.0063 -0.0010 -0.0195
( 35.22%) 0.5934* H 15 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 0.0049 0.0041 -0.0211
15. (1.98384) BD ( 1) C 13 - H 16
( 64.78%) 0.8049* C 13 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0070 0.0005 0.3453
0.0209 0.6578 0.0004 0.4454 -0.0067
0.0068 0.0043 0.0169 -0.0089 -0.0003
( 35.22%) 0.5934* H 16 s( 99.95%)p 0.00( 0.05%)
The less electronegative central atom contributes less to the C-X bond here (just 40%), which makes perfect sense given that the electronegativity of C>P. Also, the carbons contribute more (65% compared with 64% in the previous molecule) to the CH bonds, which is evidence for the hypothesis that a more electronegative central atom results in a less positively charged set of hydrogens. Both sets of data (NBO population analysis and charge analysis) suggest that electron density rests on the protons in this molecule compared with the previous one.
[S(CH3}3]+
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99412) BD ( 1) C 1 - H 2
( 64.23%) 0.8014* C 1 s( 27.24%)p 2.67( 72.71%)d 0.00( 0.05%)
0.0001 0.5219 -0.0018 0.0010 0.0906
-0.0195 0.0235 -0.0053 0.8473 0.0075
0.0012 0.0086 0.0023 0.0020 0.0193
( 35.77%) 0.5981* H 2 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0035 0.0018 0.0005 -0.0226
2. (1.98720) BD ( 1) C 1 - H 3
( 64.83%) 0.8051* C 1 s( 26.50%)p 2.77( 73.46%)d 0.00( 0.05%)
0.0000 0.5147 -0.0056 0.0006 -0.5698
-0.0118 0.5791 -0.0086 -0.2725 0.0079
-0.0143 0.0079 -0.0127 -0.0050 -0.0059
( 35.17%) 0.5931* H 3 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 0.0181 -0.0129 0.0058
3. (1.98721) BD ( 1) C 1 - H 4
( 64.83%) 0.8051* C 1 s( 26.50%)p 2.77( 73.45%)d 0.00( 0.05%)
0.0000 -0.5148 0.0056 -0.0006 0.2028
0.0145 0.7862 -0.0015 0.2739 -0.0079
-0.0028 -0.0005 -0.0151 0.0148 0.0058
( 35.17%) 0.5931* H 4 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0011 -0.0092 -0.0202 -0.0058
4. (1.98632) BD ( 1) C 1 - S 13
( 48.67%) 0.6976* C 1 s( 19.71%)p 4.07( 80.16%)d 0.01( 0.14%)
0.0003 0.4437 0.0140 -0.0033 0.7901
-0.0061 0.2127 -0.0017 -0.3633 -0.0098
0.0133 -0.0232 -0.0062 0.0230 -0.0096
( 51.33%) 0.7165* S 13 s( 16.96%)p 4.86( 82.41%)d 0.04( 0.63%)
0.0000 0.0001 0.4117 -0.0076 0.0012
0.0000 -0.7839 0.0345 0.0000 -0.2112
0.0093 0.0000 0.4039 0.0259 0.0248
-0.0599 -0.0161 0.0428 -0.0051
5. (1.98720) BD ( 1) C 5 - H 6
( 64.83%) 0.8051* C 5 s( 26.50%)p 2.77( 73.46%)d 0.00( 0.05%)
0.0000 0.5147 -0.0056 0.0006 0.7862
-0.0016 0.2040 0.0145 -0.2729 0.0079
0.0028 -0.0150 -0.0005 0.0148 -0.0059
( 35.17%) 0.5931* H 6 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 -0.0202 -0.0093 0.0058
6. (1.98721) BD ( 1) C 5 - H 7
( 64.83%) 0.8051* C 5 s( 26.50%)p 2.77( 73.45%)d 0.00( 0.05%)
0.0000 0.5147 -0.0056 0.0006 -0.5797
0.0086 0.5687 0.0118 -0.2735 0.0079
-0.0142 0.0128 -0.0079 0.0050 -0.0058
( 35.17%) 0.5931* H 7 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 0.0129 -0.0181 0.0058
7. (1.99412) BD ( 1) C 5 - H 8
( 64.23%) 0.8014* C 5 s( 27.24%)p 2.67( 72.71%)d 0.00( 0.05%)
0.0001 0.5220 -0.0018 0.0010 -0.0245
0.0052 -0.0903 0.0195 0.8473 0.0075
0.0012 -0.0023 -0.0086 -0.0020 0.0193
( 35.77%) 0.5981* H 8 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0035 -0.0005 -0.0018 -0.0226
8. (1.98631) BD ( 1) C 5 - S 13
( 48.67%) 0.6976* C 5 s( 19.71%)p 4.07( 80.16%)d 0.01( 0.14%)
-0.0003 -0.4437 -0.0140 0.0033 0.2109
-0.0016 0.7906 -0.0062 0.3633 0.0098
-0.0132 -0.0062 -0.0232 0.0230 0.0096
( 51.33%) 0.7165* S 13 s( 16.96%)p 4.86( 82.41%)d 0.04( 0.63%)
0.0000 -0.0001 -0.4117 0.0075 -0.0012
0.0000 -0.2091 0.0092 0.0000 -0.7845
0.0345 0.0000 -0.4038 -0.0259 -0.0246
-0.0160 -0.0599 0.0429 0.0051
9. (1.98721) BD ( 1) C 9 - H 10
( 64.83%) 0.8051* C 9 s( 26.50%)p 2.77( 73.45%)d 0.00( 0.05%)
0.0000 0.5147 -0.0056 0.0006 0.7822
0.0059 0.2179 -0.0133 -0.2738 0.0079
0.0115 -0.0133 -0.0071 0.0098 -0.0058
( 35.17%) 0.5931* H 10 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 -0.0221 -0.0021 0.0058
10. (1.99412) BD ( 1) C 9 - H 11
( 64.23%) 0.8014* C 9 s( 27.24%)p 2.67( 72.71%)d 0.00( 0.05%)
0.0001 0.5220 -0.0018 0.0010 -0.0658
0.0143 0.0666 -0.0143 0.8473 0.0075
-0.0023 -0.0063 0.0063 0.0000 0.0193
( 35.77%) 0.5981* H 11 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0035 -0.0013 0.0013 -0.0226
11. (1.98722) BD ( 1) C 9 - H 12
( 64.83%) 0.8051* C 9 s( 26.50%)p 2.77( 73.45%)d 0.00( 0.05%)
0.0000 0.5147 -0.0056 0.0006 -0.2163
0.0133 -0.7830 -0.0059 -0.2727 0.0079
0.0114 0.0071 0.0133 -0.0099 -0.0059
( 35.17%) 0.5931* H 12 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 0.0021 0.0222 0.0058
12. (1.98631) BD ( 1) C 9 - S 13
( 48.67%) 0.6976* C 9 s( 19.70%)p 4.07( 80.16%)d 0.01( 0.14%)
-0.0003 -0.4436 -0.0140 0.0033 0.5795
-0.0045 -0.5777 0.0045 0.3633 0.0098
0.0265 -0.0170 0.0170 -0.0001 0.0096
( 51.33%) 0.7165* S 13 s( 16.95%)p 4.86( 82.42%)d 0.04( 0.63%)
0.0000 -0.0001 -0.4117 0.0075 -0.0012
0.0000 -0.5749 0.0253 0.0000 0.5733
-0.0252 0.0000 -0.4038 -0.0259 0.0495
-0.0439 0.0438 -0.0001 0.0051
The S contributes 51% of the electrons in the S-C bonds, which makes sense. It is the 2nd most electronegative, and contributes the 2nd most electrons to the bond. The interesting non-identical hydrogens can be looked into here. The data confirms that there are two types of CH bonds present, the first involves a 35% contribution from H whereas the second involves a 36% contribution. This is evidence that the lone pair is donating electron density into the 2nd type (36%) of CH bond.
MOs of [N(CH3)4]+
A selection of MOs, starting with the HOMO and working down to a heavily bonding orbital.
Influence of fuctional groups
Two further molecules ([N(CH3)3(CH2OH)]+ & [N(CH3)3(CH2CN)]+)were optimised, a frequency analysis was carried out, and a population analysis was carried out. All relevant dats is presented below.
N(CH3)3(CH2OH)]+
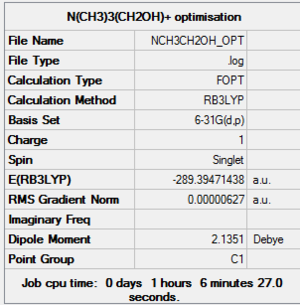
Optimisation: MEDIA:NCH3CH2OH OPT.LOG
Frequency analysis: MEDIA:NCH3CH2OH FREQ.LOG
Population analysis: MEDIA:NCH3CH2OH NBO.LOG
Item Value Threshold Converged? Maximum Force 0.000026 0.000450 YES RMS Force 0.000005 0.000300 YES Maximum Displacement 0.000797 0.001800 YES RMS Displacement 0.000214 0.001200 YES Predicted change in Energy=-1.251261D-08 Optimization completed. -- Stationary point found.
Low frequencies --- -11.8112 0.0009 0.0009 0.0013 8.1668 14.2920 Low frequencies --- 133.9649 214.0310 255.7114
NBO analysis:

Almost every atom has a different charge, tabulating would be unclear so a very large image containing the charge info has been included (above and right - click to enlarge)
The electron withdrawing OH group has an effect on the whole molecule, but especially the carbon atom adjacent to it. Every other carbon has a charge of approximately -0.490, whereas the C-OH carbon has a charge of +0.088. This is due to the highly electronegative oxygen atom. The fact that the carbon has a positive charge also affects the protons attached to it via the inductive effect. The central nitrogen is more negative in this molecule than the [N(CH3)4]+, also due to the inductive effect.
This molecule is a good illustration of why the proton of an OH group is so easily removed by base. The most positively charged atom in the whole molecule is this proton.
[N(CH3)3(CH2CN)]+

Optimisation: MEDIA:NCH3CH2CN OPT.LOG
Frequency analysis: MEDIA:NCH3CH2CN FREQ.LOG
Population analysis: MEDIA:NCH3CH2CN NBO.LOG
Item Value Threshold Converged? Maximum Force 0.000013 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.001798 0.001800 YES RMS Displacement 0.000374 0.001200 YES Predicted change in Energy=-1.261889D-08 Optimization completed. -- Stationary point found.
Low frequencies --- -7.3841 -0.0011 -0.0006 0.0005 5.9724 17.0772 Low frequencies --- 92.0183 154.5880 212.7784

The first thing to note here is that the negative charge on the central N atom is of a lower magnitude compared with any other molecule studied thus far. This is due to the electron withdrawing nature of the CN group. Unlike the molecule containing the OH group, the inductive effect in this molecule is reducing the negative charge on the central N, as it is 3 bonds away from the electron withdrawing atom (N), rather than 2.
HOMO & LUMO comparisons
| Molecule | LUMO | HOMO |
|---|---|---|
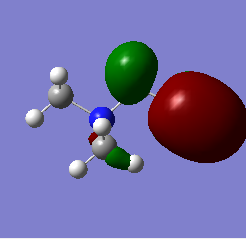
| [N(CH3)4]+ |  |
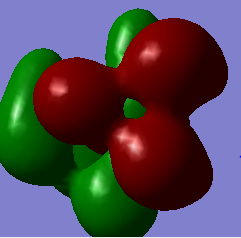
|
| N(CH3)3(CH2OH)]+ | 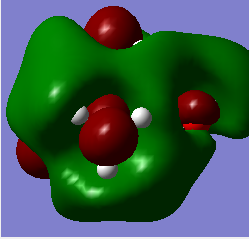 |
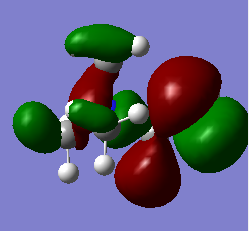
|
| N(CH3)3(CH2CN)]+ | 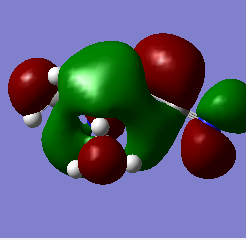 |
|
| Molecule | LUMO | HOMO |
|---|---|---|
| [N(CH3)4]+ | -0.579 | -0.133 |
| N(CH3)3(CH2OH)]+ | -0.488 | -0.125 |
| N(CH3)3(CH2CN)]+ | -0.500 | -0.182 |
The most striking picture here is the HOMO of N(CH3)3(CH2OH)]. The action of the OH group has moved a huge amount of electron density towards the OH, leaving the rest of the molecule practically bare. This is an excellent illustration of why organic chemists say OH groups are electron withdrawing, and why the carbon of -COH will so readily react with an incoming nucleophile. The -CN group also drags some electron density toward it compared with the original 'vanilla' molecule, but not nearly as much as the -OH containing one.
Another interesting thing to note is that the difference in energy between the HOMO & LUMO of the two non-vanilla molecules is reduced. This implies that it would be easier to add an electron to both of these molecules compared to the vanilla one, especially the molecule containing -CN. This is due to the electron withdrawing nature of the -CN group.
The two molecules containing -OH and -CN have a higher HOMO energy. This is simply down to the fact that those molecules contain more electrons, and so more MOs are filled, leading to a higher energy. It would therefore be easiest to remove an electron from the -OH containing molecule, hardest to remove an electron from the vanilla molecule. It would be hardest to donate an electron into the -OH molecule, since its LUMO has the highest energy value, and esiest to donate into the -CN molecule for the reverse reason.
Referenc