Rep:Mod:Js Mod1
Module 1: Organic
Introduction
Modelling using Molecular Mechanics
The Hydrogenation of Cyclopentadiene Dimer
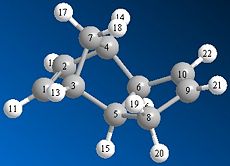
Cyclopentadiene can dimerise to form one of two products; the endo or exo dimer. Using a program such as ChemBio 3D the relative stability of each dimer can be determined and a decision as to which dimer is the kinetic and which dimer is the thermodynamic product can be made. The endo and exo dimers were drawn and their geometries optimised using the MM2 model. The results of this optimisation suggest that the exo dimer is the more thermodynamically stable product since it has a much lower total energy than the endo product; 31.89kcal/mol compared to 34.01kcal/mol respectively. The main reason for this difference in energy is due to the position of the hydrogen atoms at carbon 5 and 6 (see figure 1a. and 1b.) in relation to the bridgehead. In the exo dimer these two hydrogen atoms are orientated below the cyclopentadiene ring, on the opposite side to the bridgehead. Whereas in the endo dimer these two hydrogen atoms are orientated above the cyclopentadiene ring, on the same side as the bridgehead. This means that there is a much greater steric clash between these hydrogen atoms and the bridgehead in the endo dimer than in the exo dimer.
As can be seen in the table below the torsion energy of the endo dimer is much greater than the torsion energy of the exo dimer; 9.51kcal/mol compared to 7.63kcal/mol respectively.
We are told that the endo dimer is in fact the main dimer formed even though as mentioned above it has a much higher energy. This can be explained by considering the energy of the transition states that lead to the different dimers. The endo dimer has a lower energy transition state than the exo dimer and hence is the more kinetically stable product.
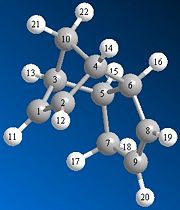
The endo dimer can undergo hydrogenation to give one of two products; dihydro derivative (3) or dihydro derivative (4) (see fig. 1e). The results of the optimisation of these two products suggest that derivative (4) is the more thermodynamically stable product since it has a much lower total energy than derivative (3); 31.16kcal/mol compared to 35.70kcal/mol respectively. As can be seen in the table below the bending energy of derivative (3) is much greater than the bending energy of derivative (4); 19.85kcal/mol compared to 14.49kcal/mol respectively.
 |
 |
 |
|
Stereochemistry of nucleophilic additions to a pyridinium ring (NAD+ analogue)
The nucleophilic addition of the grignard reagent MeMgI to the pyridinium ring (5) occurs via the mechanism shown in fig. 1f to give molecule (6).
 |
This reaction has high regio and stereoselectivity since the methyl group of the MeMgI will only add to the pyridinium ring at the 4-position. The main reason for this high selectivity is that the Mg of the MeMgI coordinates to the amide oxygen.
Molecule (5) was drawn in ChemBio 3D and the MMFF94 force field option was used to determine its energy. Slight modifications were then made to the structure of this molecule to try to minimise its energy. Fig. 1g shows the lowest energy conformation of molecule (5). The energy of this conformation is 57.48kcal/mol.
 |
The nucleophilic addition of PhNH2 to the pyridinium ring (7) occurs via the mechanism shown in fig. 1g to give molecule (8).
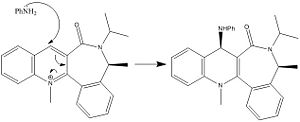
Just as for molecule (5), molecule (7) was drawn in ChemBio 3D and the MMFF94 force field option was used to determine its energy. Slight modifications were then made to the structure of this molecule to try to minimise its energy. Fig. 1i shows the lowest energy conformation of molecule (7). The energy of this conformation is 98.36kcal/mol.
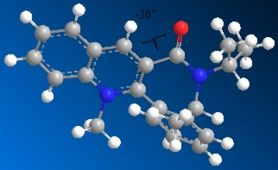 98.4513 |
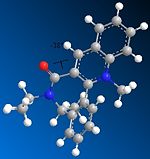 98.3634
|
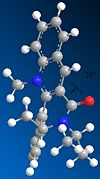 98.3744 |
Stereochemistry and reactivity of an intermediate in the synthesis of Taxol
Figure. 1k shows the mechanism that leads to the formation of molecules (9) and (10). This is an example of a atropselective oxy-Cope rearrangement. [1] What this means is that rotation about certain single bonds in each molecule is severly hindered and hence the energy needed to induce rotation about these bonds is sufficiently high enough to allow for two separate isomers to be identified.
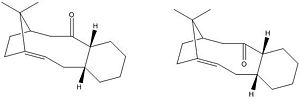
NEED TO CHECK MOLECULES AGAIN...9 SHOULD BE 10 (LOOK AT CARBONYL WITH RESPECT TO BRIDGE)

TWIST BOAT -----------MMFF94 Minimization------------
Iteration 52: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Final Energy: 66.3499 kcal/mol Calculation completed
MM2 Minimization------------
Note: All parameters used are finalized (Quality = 4).
Iteration 102: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.6548 Bend: 11.3240 Stretch-Bend: 0.3045 Torsion: 21.8117 Non-1,4 VDW: -1.3084 1,4 VDW: 13.6048 Dipole/Dipole: -0.1911
Total Energy: 48.2002 kcal/mol Calculation completed
taxol(10)

MM2 Minimization------------
Note: All parameters used are finalized (Quality = 4).
Iteration 305: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 2.7805 Bend: 16.5342 Stretch-Bend: 0.4494 Torsion: 20.6214 Non-1,4 VDW: -0.3942 1,4 VDW: 13.9542 Dipole/Dipole: 0.1818
Total Energy: 54.1272 kcal/mol Calculation completed
MMFF94 Minimization------------
Iteration 78: Minimization terminated normally because the gradient norm is less than the minimum gradient norm
Final Energy: 77.9618 kcal/mol Calculation completed
Modelling using semi empirical molecular orbital theory
Regioselective addition of dichlorocarbene

MM2 Minimization------------
Warning: Some parameters are guessed (Quality = 1).
Iteration 98: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 0.6129 Bend: 4.8462 Stretch-Bend: 0.0415 Torsion: 7.5971 Non-1,4 VDW: -1.0849 1,4 VDW: 5.7885 Dipole/Dipole: 0.1113
Total Energy: 17.9126 kcal/mol Calculation completed
Mopac Interface ------------
Model: js1007dichlorocarbenemm2
Mopac Job: AUX PM6 CHARGE=0 EF GNORM=0.100 GRAPH SHIFT=80 Finished @ RMS Gradient = 0.09750 (< 0.10000) Heat of Formation = 19.74037 Kcal/Mol
HYDROGENATED PRODUCT:
MM2 Minimization------------
Warning: Some parameters are guessed (Quality = 1).
Iteration 93: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 0.8422 Bend: 4.8812 Stretch-Bend: 0.0898 Torsion: 12.5444 Non-1,4 VDW: -1.1771 1,4 VDW: 7.5416 Dipole/Dipole: 0.0732
Total Energy: 24.7951 kcal/mol Calculation completed
Mopac Interface ------------
Model: Untitled-1
Mopac Job: AUX PM6 CHARGE=0 EF GNORM=0.100 SHIFT=80 Finished @ RMS Gradient = 0.09130 (< 0.10000) Heat of Formation = -0.36281 Kcal/Mol
Mini project
U00PLOAD JPEG
 |
anti 2b after mm2 minimal energy could find before geometry opt by scan
MM2 Minimization------------
Pi System: 12 1 2 3 11 4 6 5 7 8 10 18 Warning: Some parameters are guessed (Quality = 1).
Iteration 122: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 3.4186 Bend: 11.0621 Stretch-Bend: 0.8555 Torsion: 8.0071 Non-1,4 VDW: -5.0795 1,4 VDW: 27.5517 Dipole/Dipole: -9.3311
Total Energy: 36.4843 kcal/mol Calculation completed
syn 2b after mm2 minimal energy could find before geometry opt by scan
MM2 Minimization------------
Pi System: 12 1 2 3 11 4 6 5 7 8 10 18 Warning: Some parameters are guessed (Quality = 1).
Iteration 271: Minimization terminated normally because the gradient norm is less than the minimum gradient norm Stretch: 3.1070 Bend: 10.8083 Stretch-Bend: 0.7698 Torsion: 10.7190 Non-1,4 VDW: -7.4532 1,4 VDW: 27.1941 Dipole/Dipole: -9.7927
Total Energy: 35.3523 kcal/mol Calculation completed
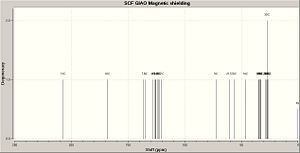
SYN 2B AFTER GEOMETRY OPT AND NMR CALC
| Chemical shift / (ppm) | Atom number |
| 207.4 | 13 |
| 168.4 | 10 |
| 136.3 | 7 |
| 134.5 | 8 |
| 128.2 | 4 |
| 126.4 | 11 |
| 125.7 | 3 |
| 125.6 | 5 |
| 123.6 | 1 |
| 123.3 | 2 |
| 122.7 | 6 |
| 120.4 | 12 |
| 72.5 | 9 |
| 60.7 | 21 |
| 56.6 | 20 |
| 46.5 | 14 |
| 34.4 | 31 |
| 34.3 | 17 |
| 34.1 | 27 |
| 33.1 | 26 |
| 32.9 | 22 |
| 28.4 | 23 |
| 28.0 | 28 |
| 27.3 | 25,30 |
| 26.6 | 24 |
| 26.0 | 29 |
 |
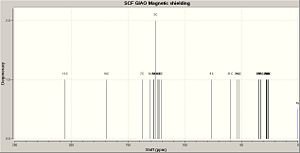
ANTI 2B AFTER GEOMETRY OPT AND NMR CALC
| Chemical shift (ppm) | Atoms |
| 205.5 | 13 |
| 169.2 | 10 |
| 137.6 | 7 |
| 130.3 | 8 |
| 127.2 | 4 |
| 126.9 | 5 |
| 125.7 | 11,3 |
| 123.8 | 1 |
| 123.1 | 2 |
| 122.6 | 12 |
| 121.0 | 6 |
| 76.2 | 9 |
| 59.7 | 21 |
| 53.6 | 20 |
| 52.2 | 14 |
| 34.7 | 26 |
| 34.3 | 17 |
| 34.2 | 31 |
| 33.1 | 27 |
| 32.6 | 22 |
| 28.2 | 23 |
| 27.7 | 25 |
| 27.5 | 28 |
| 27.4 | 30 |
| 26.4 | 24 |
| 26.2 | 29 |
 |
LINK TO SYN DOI:10042/to-3705
LINK TO ANTI DOI:10042/to-3706
References
- ↑ A. G. Schultz, L. Flood - Journal of Organic Chemistry - 1986, 51, pp838-841