Rep:Mod:JON1802
PART ONE
Jonathan Shute
Optimising BH3
Item table of converged forces and distances:
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000059 0.000300 YES
Maximum Displacement 0.000352 0.001800 YES
RMS Displacement 0.000230 0.001200 YES
Predicted change in Energy=-4.580970D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1945 -DE/DX = -0.0001 !
! R2 R(1,3) 1.1945 -DE/DX = -0.0001 !
! R3 R(1,4) 1.1945 -DE/DX = -0.0001 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
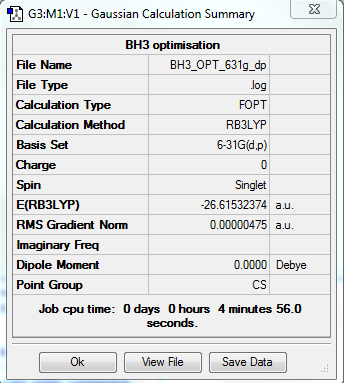
Energy is therefore: -26.4622643 a.u
Optimising BH3 second basis set
Item table of converged forces and distances:
Item Value Threshold Converged?
Maximum Force 0.000010 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.000039 0.001800 YES
RMS Displacement 0.000025 0.001200 YES
Predicted change in Energy=-5.356817D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0002 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0002 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9997 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
The Energy is -26.615(32374) a.u.
The bond length is 1.19 A
Bond Angle = 120
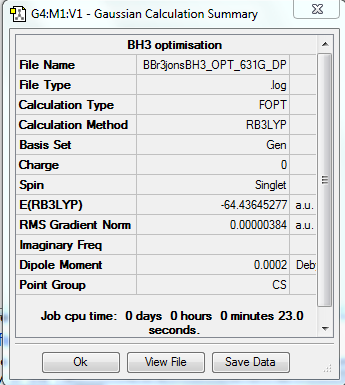
BBr3 Mixed
File:BBR3JONSBH3 OPT 631G DPsubmitwikioptomisation.txt
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000037 0.001800 YES
RMS Displacement 0.000024 0.001200 YES
Predicted change in Energy=-4.112386D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.9339 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0021 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9957 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0021 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Bond lengths are 1.93(397)A Angle = 120
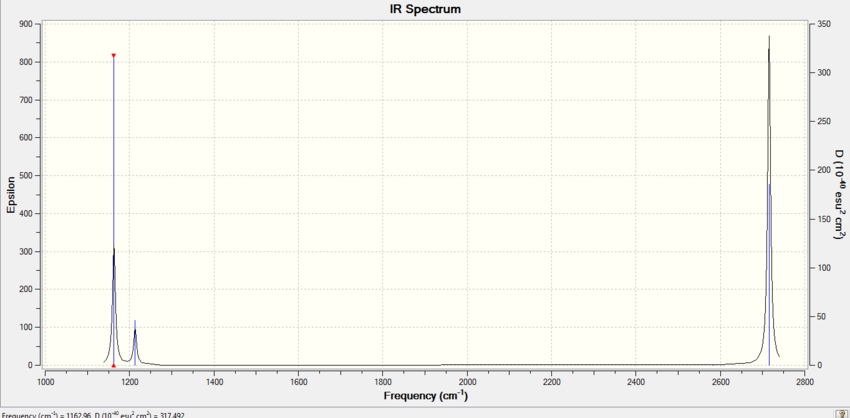
Frequency Analysis of BH3
Low frequencies --- -3.5991 -1.1355 -0.0055 1.3745 9.7046 9.7707 Low frequencies --- 1162.9825 1213.1733 1213.1760
Normal 0 false false false EN-GB X-NONE X-NONE
Vibration 4 has no net change in dipole associated with it hence it is IR inactive and isn’t seen on spectrum. Vibrations 2 and 3 absorb at exactly the same frequency so only one peak is observed for both and vibrations 5 and 6 likewise absorb at exactly the same frequency. Hence 3 peaks are observed in the spectrum. Why are they degenerate?
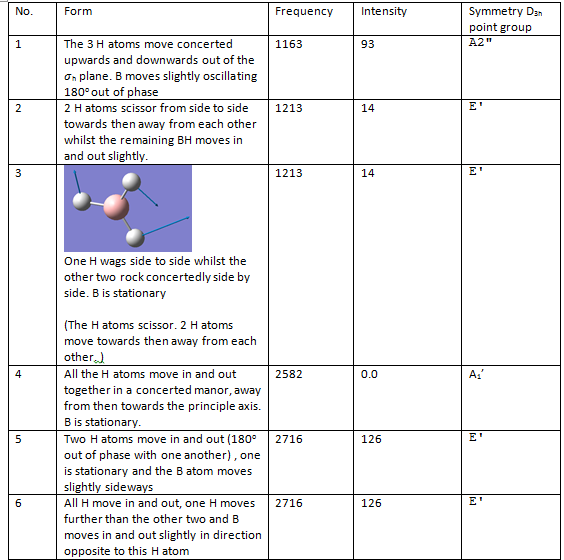
There are six catogories of vibrations [1] and broadly speaking vibration 1 is wagging, 2 is scissoring, 3 is 2 Hs rocking, 4 is symmetric stretch, 5 is assymmetric stretch and 6 is 2Hs undergoing a symmetric stretch. However, BH3 doesn't undego any twisting.
Assigning the point groups:
I redid the optimisation without nosymm:
Normal 0 false false false EN-GB X-NONE X-NONE
1 2 3
A2" E' E'
Frequencies -- 1162.9825 1213.1733 1213.1760
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9986 0.9601 0.9601
IR Inten -- 92.5497 14.0545 14.0581
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.16 0.00 0.10 0.00 -0.10 0.00 0.00
2 1 0.00 0.00 -0.57 0.00 0.08 0.00 0.81 0.00 0.00
3 1 0.00 0.00 -0.57 0.39 -0.59 0.00 0.14 -0.39 0.00
4 1 0.00 0.00 -0.57 -0.39 -0.59 0.00 0.14 0.39 0.00
4 5 6
A1' E' E'
Frequencies -- 2582.3247 2715.4994 2715.5006
Red. masses -- 1.0078 1.1273 1.1273
Frc consts -- 3.9597 4.8979 4.8979
IR Inten -- 0.0000 126.3285 126.3189
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.00 0.11 0.00 0.00 0.00 0.11 0.00
2 1 0.00 -0.58 0.00 0.02 0.00 0.00 0.00 -0.81 0.00
Normal 0 false false false EN-GB X-NONE X-NONE
BH3 MOs
DOI:10042/20684 (The population analysis)
The above orbital diagram is adapted from a second year lecture course [2].There are no significant differences between the filled calculated and LCAO MOs. The LUMO is also the same as predicted from the MO diagram however the level above this, MO 6 is somewhat misshapen. MOs 7 and 8 vary quite significantly from the predicted forms:
MO theory is useful as the orbitals involved in reactions tend to be in the HOMO-LUMO region and there is very little deviation in this area however higher MOs have a different shape than is predicted by the theory and possibly a different energy ordering if the shape deviates significantly enough.
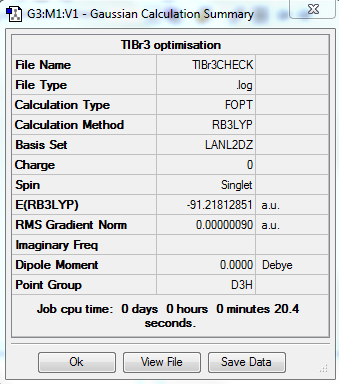
Optimising TlBr3
DOI:10042/20655 (Link to optimisation analysis file)
DOI:10042/20669 (link to frequency analysis file)
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084035D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
The optimised bond length is 2.65 A this compares to a literature value of 2.51 A which is quite close considering errors in experimental conditions for instance the measurement was taken in an aqueous solution. [3]
Structure Comparison
Bond Length Comparison
The bond lengths for BH3, BBr3, TlBr3 are 1.19, 1.93, 2.65. BH3 is electron deficient with an incomplete octet. Steric factors prevent BBr3 having such a short bond length as BH3 as H is much smaller than Br so they can fit round B even with a short bond length. Tl is a much bigger central atom than B so Br could fit around it with a shorter bond distance also there is probably less mismatch in the size of the orbitals as both Tl and Br are large atoms this might imply a stronger bond but the bonding is ionic in TlBr3 whilst it is covalent in the other molecules which may affect things too. However, there is back donation from a lone pair on Br to the B empty p orbital which strengthens and shortens the bond. This clearly is impossible with Tl. Also, because Tl is so large Br and Tl must lie quite far apart as the sum of the Van der Waals radii is greater than that of B and Br.
Further Questions
There can be a bond even when Gaussian doesn’t draw one in as it assigns a bond based on the distances between atoms. Even if the distance exceeds this, it doesn’t mean there isn’t a bonding interaction – the actual distance depends on how strong the orbital overlap is and the type of atom. It is hence hard to define a bond exactly. A bond in essence is an electronic interaction which causes two atoms to attract each other. It is different from an intramolecular bond as it is much stronger. In a covalent bond the electrons are 'shared' so attracted to both nuclei whilst an ionic bond is the electrostatic attraction caused by transfer. The bond order is the number of electrons in bonding minus antibonding orbitals. A typical covalent bond would be at least 200 kJmol-1 whilst an intramolecular H bond is about 20 kJmol-1. The strongest H bond is about 30 kJmol-1 and this energy difference distinguishes a molecular bond from an intramolecular bond.
Analysis TlBr3
| Mode# | Frequency BH3 | Frequency TlBr3 |
|---|---|---|
| 1 | 1162.96 | 46.43 |
| 2 | 1213.16 | 46.43 |
| 3 | 1213.23 | 52.14 |
| 4 | 2582.32 | 165.27 |
| 5 | 2715.48 | 210.69 |
| 6 | 2715.50 | 210.69 |
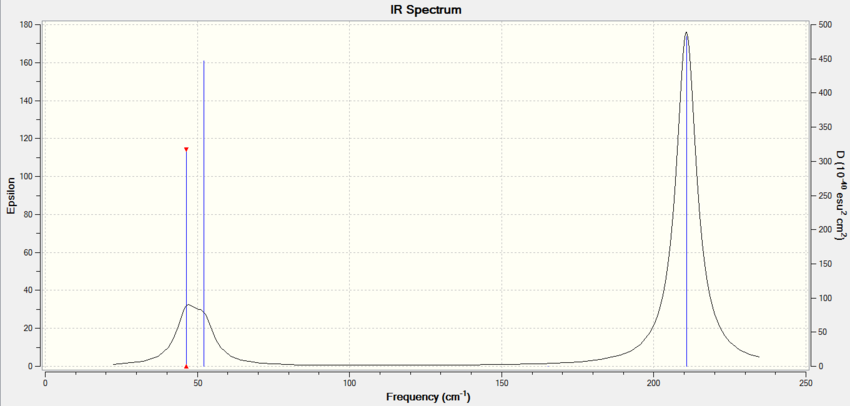
Frequency Analysis There is a large difference in the frequencies of BH3 and TlBr3 which implies there is a much stronger bond in BH3 as it vibrates at a much higher frequency. This is because Tl is a very large atom with diffuse orbitals which don't overlap well with the orbitals on Br which is also a large atom. The bond length is much larger for TlBr3 so it vibrates at a lower frequency.
The lowest real normal vibrational mode is at 46.43 cm-1
There has been a reordering of modes. Modes 2 and 3 in BH3 correspond to mode 1 and 2 in TlBr3. So the degenerate E' stretch in TlBr3 is lower in energy than A2.
The main difference between the two IR spectra is that there are 3 peaks for BH3 whilst there are only 2 for TlBr3 as the two modes at lower frequency now overlap as they have become closer together relative to each other.
Further Questions
The same method and basis set must be used as two different molecules can only be compared if they were calculated using the same method and basis set. This is because different basis sets are more accurate than others and the energy calculated varies significantly between them. However this error is assumed systematic so molecules optimised with the same method and basis set are comparable.
A frequency analysis is carried out to check whether the molecule has been optimised to a minimum in energy. If all frequencies are positive this is the case. If both are negative, it is a maximum and the optimisation would have to be repeated.
The low frequencies represent the "-6" modes in the 3N-6 vibrational modes of a molecule. They are just the motions of the center of mass of the molecule. If they are much smaller than the first vibration listed, this implies the analysis was successful. (my initial optimisation in the project for nitrobenzene failed and this was easily seen because of a very high low frequency about 200. The problem was fixed - the program had dropped the charge which was on the molecule and had optimised it as if it was neutral and the optimisation repeated.)
NH3 optimisation and population analysis
Basis set = 6-31G(d,p) Method = RB3LYP
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629731D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
--------------------------------------------------------------------------------
Low frequencies --- -30.7295 -0.0013 0.0009 0.0010 20.1705 28.2664 Low frequencies --- 1089.5535 1694.1244 1694.1856
The low frequencies are outside the range -15-15 which is expected for this level of basis set. However 1089 is a large vibration and so the low frequencies are very easily within an order of magnitude less than this.
File:JONSNH3FREQCORRECTTWO.LOG
File:JONSNH3OPTIMISATIONCORRECT.LOG
NH3 NBO
The charge range is -1.0 to +1.0
The specific NBO charge for nitrogen is -1.125 and hydrogen is +0.375
NH3BH3 Optimisation
Optimisation has converged:
Item Value Threshold Converged?
Maximum Force 0.000137 0.000450 YES
RMS Force 0.000038 0.000300 YES
Maximum Displacement 0.001020 0.001800 YES
RMS Displacement 0.000225 0.001200 YES
Predicted change in Energy=-1.139106D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0185 -DE/DX = 0.0 !
! R2 R(2,7) 1.0185 -DE/DX = 0.0 !
! R3 R(3,7) 1.0185 -DE/DX = 0.0 !
! R4 R(4,8) 1.2097 -DE/DX = 0.0 !
! R5 R(5,8) 1.2097 -DE/DX = 0.0 !
! R6 R(6,8) 1.2097 -DE/DX = 0.0 !
! R7 R(7,8) 1.6685 -DE/DX = -0.0001 !
! A1 A(1,7,2) 107.8567 -DE/DX = 0.0 !
! A2 A(1,7,3) 107.8615 -DE/DX = 0.0 !
! A3 A(1,7,8) 111.0394 -DE/DX = 0.0 !
! A4 A(2,7,3) 107.861 -DE/DX = 0.0 !
! A5 A(2,7,8) 111.0396 -DE/DX = 0.0 !
! A6 A(3,7,8) 111.0358 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.9015 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.8953 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.567 -DE/DX = 0.0001 !
! A10 A(5,8,6) 113.9013 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.5643 -DE/DX = 0.0001 !
! A12 A(6,8,7) 104.5652 -DE/DX = 0.0001 !
! D1 D(1,7,8,4) -179.9977 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -59.9951 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 60.0064 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -59.9999 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 60.0027 -DE/DX = 0.0 !
! D6 D(2,7,8,6) -179.9958 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 60.001 -DE/DX = 0.0 !
! D8 D(3,7,8,5) -179.9965 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -59.995 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Frequencies are low:
Low frequencies --- -0.0002 0.0010 0.0012 9.3484 10.9499 30.1249 Low frequencies --- 264.8031 631.3745 637.2812
NH3BH3 Association Energy
(1) E(NH3BH3) = -83.22 Au (2) E(BH3) = -26.62 Au (3) E(NH3) = -56.56 Au
The dissociation energy = (1)-[(2)+(3)] = 0.04 Au
1 hartree = 1 Au = 2625.5 kJmol-1
hence dissociation energy = 105 kJmol-1
AROMATICITY PROJECT
Jonathan Shute
Abstract
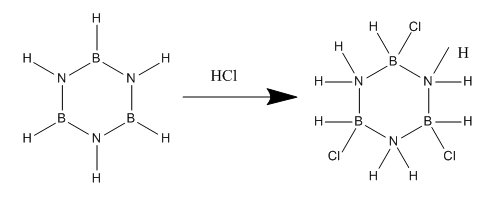
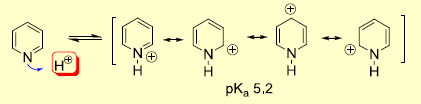
This project explores the quantum mechanical origin of the chemistry of a series of aromatic molecules: benzene, pyridinium ion, borobenzene and borazine. Why it is that borazine readily undergoes addition reactions whilst benzene will require very forcing conditions and even then undergoes aromatic substitution and not addition will be rationalised. An example of this behaviour is the following reaction scheme:
With benzene there is no reaction.
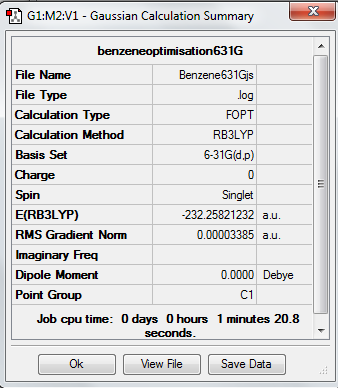
Benzene Optimisation
Item Value Threshold Converged?
Maximum Force 0.000071 0.000450 YES
RMS Force 0.000019 0.000300 YES
Maximum Displacement 0.000124 0.001800 YES
RMS Displacement 0.000049 0.001200 YES
Predicted change in Energy=-2.212397D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0 !
! R2 R(1,6) 1.3962 -DE/DX = 0.0001 !
! R3 R(1,7) 1.0864 -DE/DX = 0.0 !
! R4 R(2,3) 1.3962 -DE/DX = 0.0001 !
! R5 R(2,8) 1.0864 -DE/DX = 0.0 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0 !
! R7 R(3,9) 1.0864 -DE/DX = 0.0 !
! R8 R(4,5) 1.3962 -DE/DX = 0.0001 !
! R9 R(4,10) 1.0864 -DE/DX = 0.0 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0 !
! R11 R(5,11) 1.0864 -DE/DX = 0.0 !
! R12 R(6,12) 1.0864 -DE/DX = 0.0 !
! A1 A(2,1,6) 120.0009 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.9975 -DE/DX = 0.0 !
! A3 A(6,1,7) 120.0017 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0012 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.9967 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.0021 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.9979 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0052 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9969 -DE/DX = 0.0 !
! A10 A(3,4,5) 120.0008 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9941 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0051 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.0012 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0039 -DE/DX = 0.0 !
! A15 A(6,5,11) 119.9948 -DE/DX = 0.0 !
! A16 A(1,6,5) 119.9979 -DE/DX = 0.0 !
! A17 A(1,6,12) 120.0044 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9976 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 0.0034 -DE/DX = 0.0 !
! D2 D(6,1,2,8) -180.0015 -DE/DX = 0.0 !
! D3 D(7,1,2,3) 180.0033 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0017 -DE/DX = 0.0 !
! D5 D(2,1,6,5) 0.0015 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -180.0006 -DE/DX = 0.0 !
! D7 D(7,1,6,5) 180.0016 -DE/DX = 0.0 !
! D8 D(7,1,6,12) -0.0005 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.006 -DE/DX = 0.0 !
! D10 D(1,2,3,9) -180.0022 -DE/DX = 0.0 !
! D11 D(8,2,3,4) -180.0011 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0027 -DE/DX = 0.0 !
! D13 D(2,3,4,5) 0.0037 -DE/DX = 0.0 !
! D14 D(2,3,4,10) 180.0024 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9999 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0014 -DE/DX = 0.0 !
! D17 D(3,4,5,6) 0.0012 -DE/DX = 0.0 !
! D18 D(3,4,5,11) -180.0014 -DE/DX = 0.0 !
! D19 D(10,4,5,6) 180.0025 -DE/DX = 0.0 !
! D20 D(10,4,5,11) -0.0001 -DE/DX = 0.0 !
! D21 D(4,5,6,1) -0.0038 -DE/DX = 0.0 !
! D22 D(4,5,6,12) -180.0017 -DE/DX = 0.0 !
! D23 D(11,5,6,1) 179.9988 -DE/DX = 0.0 !
! D24 D(11,5,6,12) 0.0009 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Low frequencies --- -5.1642 -0.0006 -0.0001 0.0009 13.0466 16.7439 Low frequencies --- 414.1402 414.9607 621.1690
Low frequencies are within the approximate range of +/-15 which is reasonable considering the basis set used. (A 321G basis set might have given frequencies at 50).
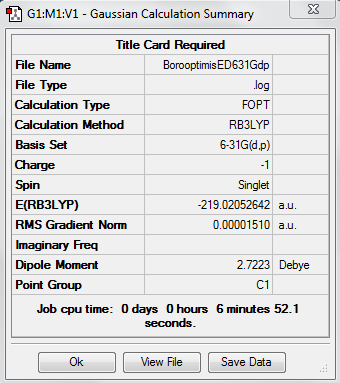
Borobenzene Reoptimisation
DOI:10042/20907 (Boro optimisation 631Gdp)
DOI:10042/20909 (Boro freq)
DOI:10042/20910 (Pop analysis)
Low frequencies --- -2.4461 -0.0008 -0.0004 -0.0004 6.7931 8.6784 Low frequencies --- 371.2420 404.5571 565.2273
Item Value Threshold Converged?
Maximum Force 0.000048 0.000450 YES
RMS Force 0.000011 0.000300 YES
Maximum Displacement 0.000526 0.001800 YES
RMS Displacement 0.000157 0.001200 YES
Predicted change in Energy=-3.252976D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3989 -DE/DX = 0.0 !
! R2 R(1,6) 1.5141 -DE/DX = 0.0 !
! R3 R(1,7) 1.0968 -DE/DX = 0.0 !
! R4 R(2,3) 1.4051 -DE/DX = 0.0 !
! R5 R(2,8) 1.0969 -DE/DX = 0.0 !
! R6 R(3,4) 1.4052 -DE/DX = 0.0 !
! R7 R(3,9) 1.0915 -DE/DX = 0.0 !
! R8 R(4,5) 1.3988 -DE/DX = 0.0 !
! R9 R(4,10) 1.0969 -DE/DX = 0.0 !
! R10 R(5,6) 1.5141 -DE/DX = 0.0 !
! R11 R(5,11) 1.0968 -DE/DX = 0.0 !
! R12 R(6,12) 1.2186 -DE/DX = 0.0 !
! A1 A(2,1,6) 120.0639 -DE/DX = 0.0 !
! A2 A(2,1,7) 116.0054 -DE/DX = 0.0 !
! A3 A(6,1,7) 123.9308 -DE/DX = 0.0 !
! A4 A(1,2,3) 122.1819 -DE/DX = 0.0 !
! A5 A(1,2,8) 120.3553 -DE/DX = 0.0 !
! A6 A(3,2,8) 117.4628 -DE/DX = 0.0 !
! A7 A(2,3,4) 120.4114 -DE/DX = 0.0 !
! A8 A(2,3,9) 119.7912 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.7974 -DE/DX = 0.0 !
! A10 A(3,4,5) 122.1782 -DE/DX = 0.0 !
! A11 A(3,4,10) 117.4619 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.3599 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.0694 -DE/DX = 0.0 !
! A14 A(4,5,11) 115.9993 -DE/DX = 0.0 !
! A15 A(6,5,11) 123.9313 -DE/DX = 0.0 !
! A16 A(1,6,5) 115.0952 -DE/DX = 0.0 !
! A17 A(1,6,12) 122.4563 -DE/DX = 0.0 !
! A18 A(5,6,12) 122.4485 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.0152 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 179.9988 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -180.0143 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0003 -DE/DX = 0.0 !
! D5 D(2,1,6,5) 0.0031 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.994 -DE/DX = 0.0 !
! D7 D(7,1,6,5) 180.0022 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.005 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0181 -DE/DX = 0.0 !
! D10 D(1,2,3,9) 180.0154 -DE/DX = 0.0 !
! D11 D(8,2,3,4) -179.9955 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0018 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0083 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0019 -DE/DX = 0.0 !
! D15 D(9,3,4,5) -180.0057 -DE/DX = 0.0 !
! D16 D(9,3,4,10) 0.0008 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.0036 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 180.0088 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -180.0102 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.0022 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.0059 -DE/DX = 0.0 !
! D22 D(4,5,6,12) -179.9969 -DE/DX = 0.0 !
! D23 D(11,5,6,1) 179.9925 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0103 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
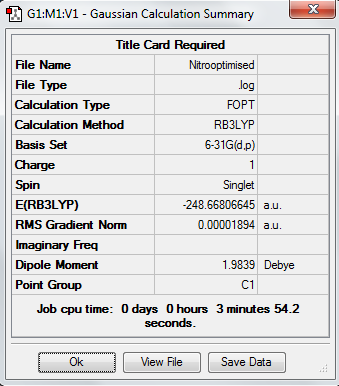
Nitro Reoptimisation
DOI:10042/20943 Optimisation 631G
DOI:10042/20945 Frequency
DOI:10042/20947 Pop analysis
Low frequencies --- -5.0089 -0.0002 0.0002 0.0004 10.1372 12.6685 Low frequencies --- 392.0778 404.2595 620.3186
Item Value Threshold Converged?
Maximum Force 0.000053 0.000450 YES
RMS Force 0.000017 0.000300 YES
Maximum Displacement 0.000518 0.001800 YES
RMS Displacement 0.000133 0.001200 YES
Predicted change in Energy=-3.700573D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3837 -DE/DX = 0.0 !
! R2 R(1,6) 1.3524 -DE/DX = 0.0 !
! R3 R(1,7) 1.0832 -DE/DX = 0.0 !
! R4 R(2,3) 1.3988 -DE/DX = 0.0 !
! R5 R(2,8) 1.0835 -DE/DX = 0.0 !
! R6 R(3,4) 1.3987 -DE/DX = 0.0 !
! R7 R(3,9) 1.0852 -DE/DX = 0.0 !
! R8 R(4,5) 1.3838 -DE/DX = 0.0 !
! R9 R(4,10) 1.0835 -DE/DX = 0.0 !
! R10 R(5,6) 1.3524 -DE/DX = 0.0 !
! R11 R(5,11) 1.0832 -DE/DX = 0.0 !
! R12 R(6,12) 1.0169 -DE/DX = 0.0 !
! A1 A(2,1,6) 119.2334 -DE/DX = 0.0 !
! A2 A(2,1,7) 123.9404 -DE/DX = 0.0 !
! A3 A(6,1,7) 116.8262 -DE/DX = 0.0 !
! A4 A(1,2,3) 119.0866 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.4245 -DE/DX = 0.0001 !
! A6 A(3,2,8) 121.489 -DE/DX = 0.0 !
! A7 A(2,3,4) 120.0576 -DE/DX = 0.0 !
! A8 A(2,3,9) 119.9731 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9693 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.0802 -DE/DX = 0.0 !
! A11 A(3,4,10) 121.4996 -DE/DX = 0.0 !
! A12 A(5,4,10) 119.4202 -DE/DX = 0.0001 !
! A13 A(4,5,6) 119.2377 -DE/DX = 0.0 !
! A14 A(4,5,11) 123.94 -DE/DX = 0.0 !
! A15 A(6,5,11) 116.8223 -DE/DX = 0.0 !
! A16 A(1,6,5) 123.3046 -DE/DX = 0.0 !
! A17 A(1,6,12) 118.345 -DE/DX = 0.0 !
! A18 A(5,6,12) 118.3503 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.014 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 179.9965 -DE/DX = 0.0 !
! D3 D(7,1,2,3) 179.9909 -DE/DX = 0.0 !
! D4 D(7,1,2,8) 0.0014 -DE/DX = 0.0 !
! D5 D(2,1,6,5) -0.0018 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.9825 -DE/DX = 0.0 !
! D7 D(7,1,6,5) 179.9937 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.013 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0147 -DE/DX = 0.0 !
! D10 D(1,2,3,9) -179.978 -DE/DX = 0.0 !
! D11 D(8,2,3,4) -179.9961 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0112 -DE/DX = 0.0 !
! D13 D(2,3,4,5) 0.0001 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -179.9929 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9928 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0002 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.0156 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 179.9933 -DE/DX = 0.0 !
! D19 D(10,4,5,6) 179.9775 -DE/DX = 0.0 !
! D20 D(10,4,5,11) -0.0136 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.0169 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 179.9976 -DE/DX = 0.0 !
! D23 D(11,5,6,1) -179.9914 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0107 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
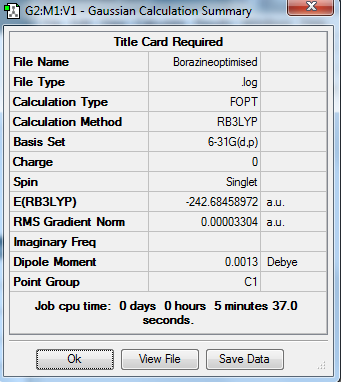
Borazine Optimisation and analysis
DOI:10042/20957 (Borazin population analysis)
DOI:10042/20789 (Borazine optimisation file)
DOI:10042/20790 (Borazine frequency analysis file)
Low frequencies --- -17.8262 -12.6392 0.0003 0.0007 0.0008 13.0244 Low frequencies --- 288.8763 289.8956 404.5100
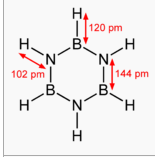
I compared my generated bond lengths with those on the borazine wikipedia
page. [4]:
My results were very close confirming my optimisation was successful: 101 pm (NH), 119.5 (BH),143 (BN)
Item Value Threshold Converged?
Maximum Force 0.000043 0.000450 YES
RMS Force 0.000016 0.000300 YES
Maximum Displacement 0.000592 0.001800 YES
RMS Displacement 0.000208 0.001200 YES
Predicted change in Energy=-7.802477D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4305 -DE/DX = 0.0 !
! R2 R(1,6) 1.4306 -DE/DX = 0.0 !
! R3 R(1,7) 1.1951 -DE/DX = 0.0 !
! R4 R(2,3) 1.4307 -DE/DX = 0.0 !
! R5 R(2,8) 1.0097 -DE/DX = 0.0 !
! R6 R(3,4) 1.4306 -DE/DX = 0.0 !
! R7 R(3,9) 1.1951 -DE/DX = 0.0 !
! R8 R(4,5) 1.4306 -DE/DX = 0.0 !
! R9 R(4,10) 1.0097 -DE/DX = 0.0 !
! R10 R(5,6) 1.4306 -DE/DX = 0.0 !
! R11 R(5,11) 1.1952 -DE/DX = 0.0 !
! R12 R(6,12) 1.0097 -DE/DX = 0.0 !
! A1 A(2,1,6) 117.113 -DE/DX = 0.0 !
! A2 A(2,1,7) 121.4427 -DE/DX = 0.0 !
! A3 A(6,1,7) 121.4443 -DE/DX = 0.0 !
! A4 A(1,2,3) 122.8848 -DE/DX = 0.0 !
! A5 A(1,2,8) 118.5804 -DE/DX = 0.0 !
! A6 A(3,2,8) 118.5348 -DE/DX = 0.0 !
! A7 A(2,3,4) 117.1244 -DE/DX = 0.0 !
! A8 A(2,3,9) 121.4322 -DE/DX = 0.0 !
! A9 A(4,3,9) 121.4434 -DE/DX = 0.0 !
! A10 A(3,4,5) 122.8678 -DE/DX = 0.0 !
! A11 A(3,4,10) 118.575 -DE/DX = 0.0 !
! A12 A(5,4,10) 118.5572 -DE/DX = 0.0 !
! A13 A(4,5,6) 117.1299 -DE/DX = 0.0 !
! A14 A(4,5,11) 121.4471 -DE/DX = 0.0 !
! A15 A(6,5,11) 121.423 -DE/DX = 0.0 !
! A16 A(1,6,5) 122.8802 -DE/DX = 0.0 !
! A17 A(1,6,12) 118.5675 -DE/DX = 0.0 !
! A18 A(5,6,12) 118.5524 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.023 -DE/DX = 0.0 !
! D2 D(6,1,2,8) -180.034 -DE/DX = 0.0 !
! D3 D(7,1,2,3) 180.018 -DE/DX = 0.0 !
! D4 D(7,1,2,8) 0.007 -DE/DX = 0.0 !
! D5 D(2,1,6,5) 0.0225 -DE/DX = 0.0 !
! D6 D(2,1,6,12) 180.0129 -DE/DX = 0.0 !
! D7 D(7,1,6,5) -180.0185 -DE/DX = 0.0 !
! D8 D(7,1,6,12) -0.0281 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.018 -DE/DX = 0.0 !
! D10 D(1,2,3,9) 180.0128 -DE/DX = 0.0 !
! D11 D(8,2,3,4) -179.971 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0238 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.012 -DE/DX = 0.0 !
! D14 D(2,3,4,10) 179.9372 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9932 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0576 -DE/DX = 0.0 !
! D17 D(3,4,5,6) 0.0115 -DE/DX = 0.0 !
! D18 D(3,4,5,11) -179.9958 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -179.9377 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.055 -DE/DX = 0.0 !
! D21 D(4,5,6,1) -0.0169 -DE/DX = 0.0 !
! D22 D(4,5,6,12) -180.0074 -DE/DX = 0.0 !
! D23 D(11,5,6,1) -180.0097 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0001 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Benzene Population Analysis

Discussion
(Note that I made a correction to level 14, if this doesn't show click on table and click on current version) This table represents benzene's MO diagram and it includes all the information in an MO diagram. It shows the calculated MOs and the LCAO interpretations of the MOs. The table is designed so as to highlight the similarities between the MOs. Each row contains MOs which have similar symmetry. (The actual symmetry labels do differ because of the difference in symmetry between pi and sigma hence the top left orbital is totally symmetric A1g whilst orbital with energy number 12 is A2u)
The orbitals are numbered according to their energy levels in an energy level diagram, with degenerate E orbitals having the same number. The HOMO is level 14 pi2. (The final column contains orbitals which don't fit in with the symmetry groups in the table)
The first column contains all the orbitals formed by the 1s on C. They are far lower in energy than any of the other orbitals as they contain no valence orbital contributions. The second column contains all the orbitals formed between overlap between H 1s and C 2s Third column contains all the orbitals formed from overlap between the pz orbitals. They contain only pi type side on bonding. These are the key orbitals with regards aromaticity and their symmetry is: (A2u, E1g, E2u, B2g) The first 3 in the fourth column are formed from overlap between py and H 1s. The final one is formed from overlap of px.
The energy ordering is as expected until energy level 8 where the overlap between p and s orbitals is lower than the last orbital formed from overlap with s and s orbitals. This is unexpected insofar as s orbitals are lower in energy than p orbitals as the electron spends more time on average closer to the nucleus. For the s-s overlap orbital (level 9) there is good overlap between the H's and the C's strengthening the bond between them, however the C s orbitals form a destructive overlap and there is node through every C-C bond (which probably weakens the C-C bonds rather than strengthening them)
This contrasts sharply with the case for the p-s overlap orbital (level 8) where there is a strong sigma type interaction between the H s orbitals and the C p orbital and a weaker (as sigma overlap is stronger than pi) pi type constructive overlap between p's on adjacent C's. Furthermore, there is a pool of constructive interactions in the center of the ring. The fact level 8 contains higher energy orbitals is overcompensated for by their much better overlap in this energy level.
The pi1 a2u orbital is separated by 2 energy levels from the other pz overlap orbitals. This is due to there being no nodal planes in this orbital. As a general rule, the more nodal planes, the higher in energy the orbital thus going down column one, the energy increases whilst the nodal planes go: 0, 1, 2,3.
Further Questions
The orbitals relate to aromaticity as some orbitals (e.g. pi1) are formed from 6 atomic orbitals, one on each carbon atom. This means that the electrons in pi1 are delocalised all over the ring system. All electrons in the orbitals pi1 to pi6 cover all six C atoms so the electrons in these orbitals are shared by all six C atoms equally making the 'partial double bonds'.
For aromaticity to occur, the molecule must obey Huckel's rule and there must be a cyclic planar array of p orbitals. The significance of there being a cyclic array of p orbitals is that electrons can 'travel around' the ring which lowers the energy of the orbital.
Benzene is stabilised by delocalisation, it is a hybrid of two resonance forms with alternating single and double bonds and is hence more stable than either of these individually. This gives rise to effectively six 1 and a half bonds as usually there are 2xAOs giving rise to 2 MOs, but as each pi orbital overlaps with each of its neighbours, there are 12xAOs giving rise to 12xMOs across the whole system so in effect "half" bonds are possible. Benzene is also stabilised by being a ring which allows and extra interaction between the two orbitals which would have been separated at opposite ends of corresponding chain. This leads to stabilisation if there are 4n+2 electrons.
The greatest contribution to aromatic stability is the pi1 which is so low in energy, 2 sigma orbitals come between it and pi2. In accordance with Huckel's rule, all the bonding pi orbitals must be filled for aromatic stability which achieves a closed shell of pi orbitals so there has to be 4n+2 electrons.
MO Comparison

Pi1 is essentially similar for all the molecules. However, pyridinium's pi 1 bulges outwards as it nears the N atom as the electron density is skewed towards the more electronegative atom as highlighted by the side on view. In borozene, the electron ring is slightly distorted so as to extend less far out towards the electropostive boron atom. In borazine, when viewed from above the electron cloud has slightly angular sides and is quite round as it extends a little further over the N atoms and less far out over the B atoms.
These differences are magnified in the seventh MO. This may be because the seventh MO is very bonding and so the coefficient of the orbitals on B is smaller corresponding to this extra bonding character. The H attached to B's orbital has a 1s which doesn't seem to be part of the MO (although maybe it is and the electron density has just migrated away). Being attached to B means it is quite negative (and so maybe too high energy to be part of this combination). The electron density hardly stretches over the B atom. In pyridinium, the electron cloud migrates towards the half of the ring near the N atom. In borazine, the electron cloud is completely triangular and even has dimples near the B atoms.
Antibonding MO number 24 has a break in the ring of electron density in borobenzene. Likewise pyridinium contains breaks in the ring of electron density and a loss of symmetry. Borazine remains C3 symmetric but there is much more larger orbital coefficient around the N-H H atoms than the B-H H atoms. In the NBO anlaysis below, the N-H H atoms have the more charge than the B-H H atoms however this orbital is antibonding and so unpopulated.Larger orbital coefficients around the more electropositive atoms is expected in the antibonding orbitals.
However some electron density does pool around the electropositive B atom in bonding orbitals for instance MO 11 has much more electron density on B atoms than N:
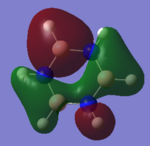
However, overall there is on average more electron density over N than B in borazine.
The Effect on the full MO diagram
Substituents cause a reduction in symmetry and so a loss of degeneracy of the energy levels. Borazine has highest symmetry element C3 whilst pyridinium ion is reduced to C2. This has the affect of altering the point group of the molecule and the symmetry labels of the orbitals may change and hence orbitals which can interact might change. However the crucial orbitals involved with aromaticity remain similar -the pi orbitals.
The sigma orbitals do change quite significantly - for borobenzene, the first six contain but 1 degenerate pair and the sixth one is much higher in energy (than in benzene). This is due to there being a 1s on B which is higher in energy than C due again to electronegativity difference.
The aromatic stabilisation varies between molecules but comparing pi1 wouldn't give a fair comparison of this for instance, pi1 is sunk very low in energy in pyridinium which is due to it being positively charged (and N being electronegative) rather than to do with aromatic stability. The HOMO is now MO 29! A possible extension to the project could be to calculate the aromatic stability by comparing the energy of cyclohexene to cyclohexane and adding on 3 times the difference. This number would be compared with the energy of benzene and the difference is the aromatic stability (and stability from delocalisation).
NBO Analysis
Note, the range is 0.2 to -0.2. Red is electron rich and green is electron poor.
Summary
1) Borobenzene has C atoms with slightly more electron density than in benzene. Although B is more electropositive than C, there is a negative charge dissociated over the molecule. B is slightly less electron rich than the C atoms. Each H atom has gained some electron density, each has the same electron density as every other. Overall, the negative charge has been delocalised very evenly across the molecule.
2) Pyridinium ion is positive but the charge is delocalised less evenly. The N atom is electronegative and so high in electron density (relative to C in benzene). The H atoms are more electron deficient, each with the same electron density. The C para to N is electron poor, the 2 Cs in ortho postion are the same electron density (as in benzene) whilst the 2 Cs meta to the N atom are very electron deficient. The following diagram taken from second year lecture course [5]
implies that the majority of positive charge should be located in the meta and para positions. This is born out in my NBO analysis which also shows that the resonance forms are weighted such that those in meta position are more significant resonance forms.
3) Borazine is of course a neutral molecule and the negative charge pools around the electronegative N atoms whilst the B atoms have very little electron density. The H atoms bonded to B have much more electron density than in benzene, and the H atoms bonded to N have much less. This is expected as the electronegative N atom 'sucks' electron density inductively away from the H atom whilst B is more electropositive than H and so the electron density is drawn towards the H atom.
Conclusion
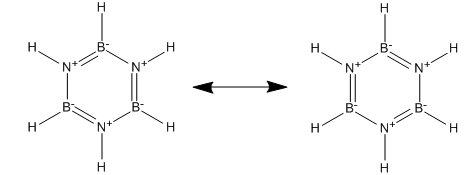
Benzene is aromatic due to the overlap of 6 p orbitals containing 1 electron each whilst borazine has overlap between the filled p orbital lone pairs on N with the empty p orbitals on boron. This amounts to electron donation from the N to the B and so borazine can be represented as:
However this is misleading as it implies the N is more positive than the B. My NBO analysis shows this not to be the case and the majority of MOs reveal that the charge is located more on N than on B. It would also not explain the product which is formed from the reaction of broazine and HCl. The nucleophilic Cl- adds to the B atoms and not the N, further evidence that the B atom is less negative than N. The NBO revealed the uneven charge distribution between B and N unlike in benzene where all C's have the same slight charge. This explains the kinetic instability of borazine as the charges attract attacking species.
The aromatic stability of borazine is also much less than benzene so it is more 'willing' to give it up in an addition reaction. This is due to the energy mismatch between B and N causing a weaker overlap between the p orbitals. The pyridinium ion should be reactive towards SNAr due to the electron withdrawing effects of N and the positive charge causing the ring to be very electron deficient. The charge distribution is also uneven. Pyridinium has no more aromatic stability than pyridine which has a stabilitiy of 117 kJmol-1 compared to 150 for benzene [6]
References
- ↑ http://www.pslc.ws/macrog/irabs.htm
- ↑ Page 2 from a tutorial sheet by Dr Hunt http://www.huntresearchgroup.org.uk/teaching/teaching_comp_lab_year3/Tut_MO_diagram_BH3.pdf
- ↑ www.actachemscand.dk/pdf/acta_vol_36a_p0125-0135.pdf Julius Glaser, Georg Johnson. Acta.Chemica.Scandinavica A 36 1982 page 125-135
- ↑ http://en.wikipedia.org/wiki/Borazine.
- ↑ Heteroaromatics lecture 6 slide 6 by Dr Spivey http://www.ch.ic.ac.uk/spivey/teaching/org2heteroaromatics/lecture61112.pdf.
- ↑ http://en.wikipedia.org/wiki/Pyridine