Rep:Mod:JFG01347171
Gaussian/Gaussview molecules
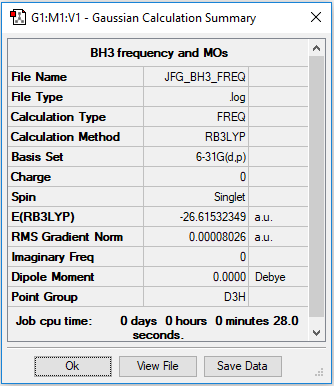
Borane
Optimised using B3LYP/6-31G
Item Value Threshold Converged?
Maximum Force 0.000161 0.000450 YES
RMS Force 0.000080 0.000300 YES
Maximum Displacement 0.000632 0.001800 YES
RMS Displacement 0.000316 0.001200 YES
Predicted change in Energy=-1.521752D-07
Optimization completed.
-- Stationary point found.
Low frequencies --- -0.1189 -0.0049 -0.0006 42.1904 42.1905 43.2823
Low frequencies --- 1163.5873 1213.5509 1213.5511
Optimised Borane molecule |
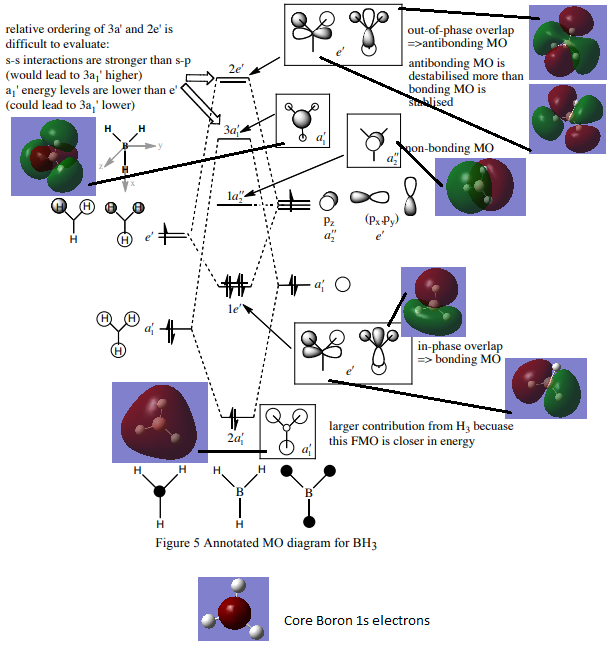
Basis of diagram taken from http://www.huntresearchgroup.org.uk/teaching/teaching_comp_lab_year2a/Tut_MO_diagram_BH3.pdf. Accessed 03/05.19.
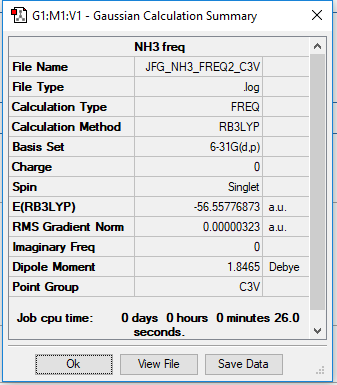
Ammonia
Optimised using B3LYP/6-31G
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000013 0.001800 YES
RMS Displacement 0.000007 0.001200 YES
Predicted change in Energy=-1.131315D-10
Optimization completed.
-- Stationary point found.
Low frequencies --- -0.0129 -0.0033 -0.0025 7.0747 8.1044 8.1047
Low frequencies --- 1089.3849 1693.9369 1693.9369
Optimised Ammonia molecule |
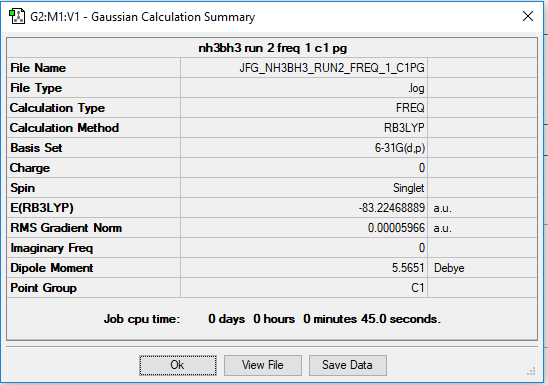
NH3BH3
Optimised using B3LYP/6-31G
Optimised NH3BH3 molecule |
Calculation of the energy of the NB bond. Calculation shows the bond is weaker than usual NB bond. -83.22468889 -(-26.61532349 - 56.55776873) = -0.05159667 a.u. -0.05159667*2625.5 = -135 kJ/mol
Good calculation of the BH3, NH3 and NH3BH3 structures and inclusion of all the required structure information in the report. The calculation of the association energy is correct but could be improved by referencing your literature value. Sadly, the section is largely incomplete with a lot of content and answers missing (e.g. NI3 calculation, the vibrational analysis of BH3) Smf115 (talk) 21:42, 13 May 2019 (BST)
Lewis acids and bases project
Diborane
Optimised using B3LYP/6-31G
Item Value Threshold Converged?
Maximum Force 0.000143 0.000450 YES
RMS Force 0.000038 0.000300 YES
Maximum Displacement 0.001510 0.001800 YES
RMS Displacement 0.000443 0.001200 YES
Predicted change in Energy=-1.460513D-07
Optimization completed.
-- Stationary point found.
Link to .log file for diborane complex
Optimised diborane complex |
Al2Br2Cl4
Of the five isomers of this compound, the bromine bridging complex and the trans bromine complex were analysed.
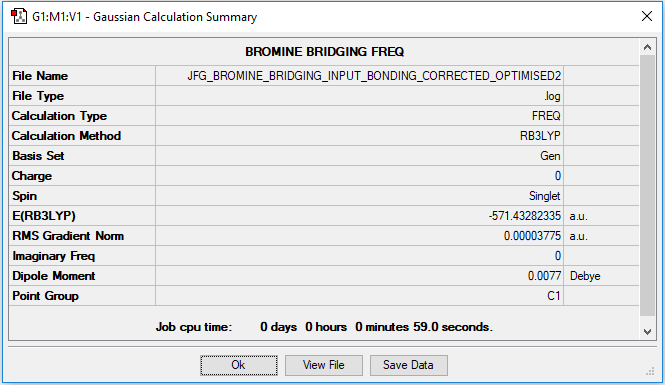
Bromines bridging: Al2Br2Cl4
Item Value Threshold Converged?
Maximum Force 0.000081 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.001895 0.001800 NO
RMS Displacement 0.000609 0.001200 YES
Predicted change in Energy=-2.331099D-07
Low frequencies --- -1.3436 -1.2172 -0.0004 0.0007 0.0014 1.1392
Low frequencies --- 16.0860 62.4928 84.8122
Link to .log file for bromine bridging complex
Optimised Bromine bridging complex |
Bromines trans: Al2Br2Cl4
Item Value Threshold Converged?
Maximum Force 0.000096 0.000450 YES
RMS Force 0.000027 0.000300 YES
Maximum Displacement 0.001438 0.001800 YES
RMS Displacement 0.000447 0.001200 YES
Predicted change in Energy=-9.489518D-08
Optimization completed.
-- Stationary point found.
Low frequencies --- -3.1045 -0.8260 -0.0007 0.0005 0.0011 1.9972
Low frequencies --- 18.8120 47.6569 71.2524
Optimised Bromine trans complex |
By taking the difference between the energies of the optimised structures, the relative stability of the trans bromine compound is: −0.0051029 a.u. = 13 kJ/mol.
(-571.43792625--571.43282335 = −0.0051029) (-0.0051029*2625.5) = −13.39766395 kJ/mol)
As such the compound with the bromines bridging is clearly the more stable of the isomers, since its computed energy is lower.
Dissociation energy of Bromine trans complex
Item Value Threshold Converged?
Maximum Force 0.000100 0.000450 YES
RMS Force 0.000052 0.000300 YES
Maximum Displacement 0.001493 0.001800 YES
RMS Displacement 0.000927 0.001200 YES
Predicted change in Energy=-9.847385D-08
Optimization completed.
-- Stationary point found.
Low frequencies --- -0.0011 0.0006 0.0015 3.5705 5.9015 6.3396
Low frequencies --- 119.8512 132.8069 182.7664
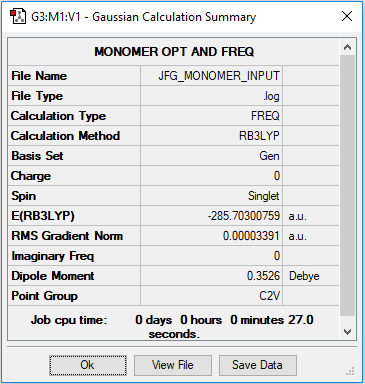
Link to .log file for monomer compound
Monomer AlBrCl2 |
By optimising a molecule of AlBrCl2, doubling its calculated energy and comparing to the energy of the trans bromine compound, the energy of dissociation can be determined.
-285.70300759*2 = -571.4060152 -571.4060152--571.43792625 = 0.03191105 a.u. 0.03191105*2625.5= 84 kJ/mol
The calculated value of 84 kJ/mol indicates that the trans bromine complex is more stable than the two monomers, respectively.
Good attempt at the structures and you have successfully included a pseudopotential for the calculation. However, the pseudopotential should have only been used on Br resulting in the wrong energies. To improve you could have considered the symmetries too of the isomers calculated, the section asking to present all five isomers and their point groups is missing which may have helped you consider this detail Smf115 (talk) 21:43, 13 May 2019 (BST)
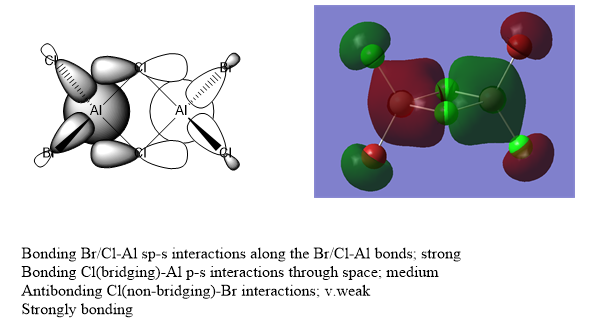
MOs of bromine trans comound
Nice selection of a range of MOs (but should have labelled which ones they were!). You have evaluated the overall character of the MOs well and in general, your LCAO diagrams are well constructed and correct apart from the missing bridging Cl orbitals on the bottom one. While you’ve highlighted some of the main interactions, the analysis could have included a few more details/annotations e.g. about the nodal planes, and clearly been labelled on the diagrams. Smf115 (talk) 21:46, 13 May 2019 (BST)
Overall, a decent report and lab which is let down by missing some of the sections. Smf115 (talk) 21:46, 13 May 2019 (BST)