Rep:Mod:Ginger1
Modelling using Molecular Mechanics
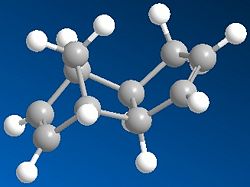
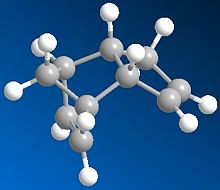
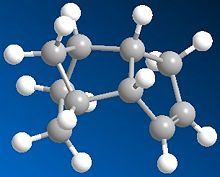
The Hydrogenation of Cyclopentadiene Dimer
Cyclopentadiene can dimerise to form two compounds an endo form (2) and an exo form (1), however the product formed is the endo (2) rather than exo (1). When this is then hydrogenated it gives both 3 and 4. The results below are after the using MM2 force field option in ChemBio3D.
 |
 |
 |
 |
| Energy kcal/mol | 1 | 2 | 3 | 4 |
|---|---|---|---|---|
| Stretch | 1.2854 | 1.2548 | 1.2321 | 1.0946 |
| Bend | 20.5764 | 20.8588 | 18.9674 | 14.5033 |
| Torsion | 7.6436 | 9.5039 | 12.1494 | 12.4936 |
| van der Waals | 4.2406 | 4.2963 | 5.7329 | 4.5230 |
| Hydrogen Bonds | -1.4019 | -1.5119 | -1.5555 | -1.0498 |
| Total Energy | 31.8821 | 34.0129 | 35.9309 | 31.1550 |
As can be seen of the two cyclopentadiene dimers the thermodynamic product is the exo product as it has a lower total energy. However the endo product is the dimer produced, thus the endo product is the kinetic product and therefore the dimerisation of cyclopentadiene is kinetically controlled. The endo product has a higher energy than the exo product as you can see that the endo product is less planar than the exo product; this can be shown by the higher torsion energy in the endo product which is a result of increased steric hinderance in the molecule.
For the 3/4 compound pair, when the energy is observed in closer detail it can be seen that for stretching 3~4, torsion 3~4 and van der Waals 3>4 but only by ~ 1kcal.mol. For the bending energy however 3>>4 and has a large difference of ~4.5kcal/mol. This therefore means that 4 is the thermodynamic product as it has a lower total energy and 3 is the kinetic product. The large difference in the bending energy can be explained by ring strain; when the sp2 carbons in the double bonds are observed the ideal angle will be 120o, for the five membered ring double bond the angle is 113o and for the six membered ring double bond the angle is 108o. Therefore when 4 is produced the most strain is released and the double bond with the least hinderance is still present so the most energy is released when compared to 3. It can be concluded from these calculation that 4 is most likely to be the major product due to the large energy difference between the kinetic and thermodyanic product.
Stereochemistry of Nucleophilic additions to a pyridinium ring (NAD+ analogue)
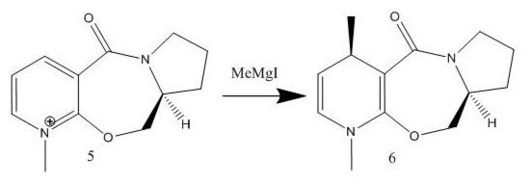
Alkylation of prolinol
This reaction involves alkyating a pyridine ring in the 4-position by using a derivative of prolinol 5 which reacts with methyl magnesium iodide. A reaction scheme can be viewed below and a jmol of compound 5 can also be viewed below.
The energy for compound 5 is 44.2807 kcal/mol, this then minimises to 39.9863 kcal/mol for compound 6. When different starting points are used e.g. carbonyl oxygen above and below the plane, methyl groups above and below the plane etc. the same optimised structure is produced and gives the same energy with the CO bond slightly above the aromatic ring as can be seen in the jmol. However if the starting point is drastically different it can produce a much higher energy of ~150 kcal/mol and will stay at this value showing that the program can minimise to local minima giving misleading results. When the modelling was attempted with MeMgI the program did not work as Mg is not supported in Chem3D.
The mechanism for this reaction involves the Mg coordinating to the carbonyl oxygen and the methyl group coordinating to the aromatic ring via a six membered transition state ring, therefore as the carbonyl oxygen is slightly angled above the plane (~12o) it means the methyl group is also angled above the plane in the transition state and therefore the methyl group attaches above the plane, resulting in product six being the stereoproduct.[1]
 |
 |
 |
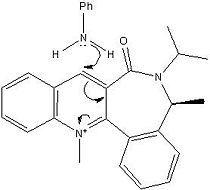
Reaction with aniline
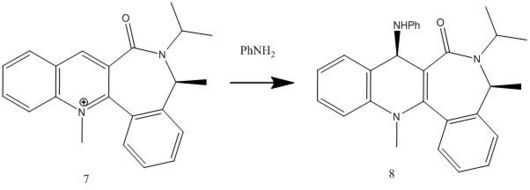
This reaction involves reacting aniline with the pyridinium ring of 7 to form 8 which acts to transfer the NHPhenyl group to electrophiles in a reverse of the reaction by which it was formed. A reaction scheme can be viewed below and a jmol of compound 7 can also be viewed below.
The energy for compound 7 is 62.1276 kcal/mol, this then minimises to 60.8028 kcal/mol for compound 8. Like in compound 5 when different starting points are used e.g. carbonyl oxygen above and below the plane, methyl groups above and below the plane etc. the same optimised structure is produced and gives the same energy with the CO bond slightly above the aromatic ring as can be seen in the jmol.
Similar to compound 5 the carbonyl oxygen is slightly angled above the plane (~23o) as can be seen from the jmol and therefore the lone pair from the nitrogen on the NH2Ph group attacks the carbon from the opposite face to the electron rich carbonyl oxygen thus avoiding electron repulsion, the new secondary amine group is therefore now on the same face as the methyl group on the 7 membered ring (the opposite face of the carbonyl oxygen).
 |
 |
 |
These simple models could be improved by including MO interactions so that orbital mixing is included and therefore diffenet types of bonds instead of only having repulsions, torsion, sterics etc. as there is only so much that models based on these simple parameters can do.
Stereochemistry and Reactivity of an Intermediate in the Synthesis of Taxol
 |
 |
During the synthesis of Taxol there is a key intermediate 9 or 10, the difference between these two intermidates is that in 9 the carbonyl oxygen is pointing up and in 10 the carbonyl oxygen is pointing down.
After several MM2 optimisations and manual editing of the structure in ChemBio3D intermediate 9 forms a twist-boat conformer with an energy of 54.1265 kcal/mol and intermediate 10 forms a chair conformer with an energy of 44.3052 kcal/mol. The most stable isomer is therefore intermediate 10 with a much lower energy than 9 by ~10kcal/mol. The difference in energy is primarily down to steric hinderance as the carbonyl oxygen in 9 points up causing the two rings point down causing unfavourable interaction whereas in 10 the carbonyl oxygen points down and therefore the two rings do not interacte with each other causing the molecule to be more stable.
This alkene reacts abnormally slowly due to the fact that it is a hyperstable alkene. This means that it is more stable then the parent alkene and is because of a negative olefin strain energy (difference between the energy of an olefin and that of its parent hydrocarbon). The reason that this is more stable is due to the location of the bridging and this results in a decrease in reactivity.[2]
Modelling Using Semi-empirical Molecular Orbital Theory
Regioselective Addition of Dichlorocarbene
Part 1

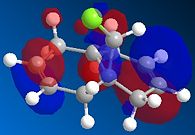
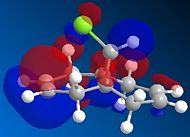
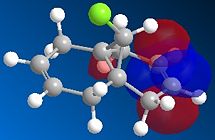
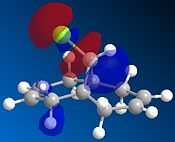
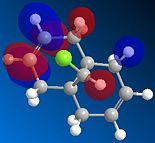
Compound 12 reacts with electrophilic reagents such as dichlorocarbene or peracid. In order to model reactions like this the geometry of compound 12 must be predicted and the energy of the orbitals calculated. Initially compound 12 was minimised using MM2 resulting in an energy of 17.8975 kcal/mol and then orbitals were calculated as seen below.
The HOMOs and LUMOs below show that the MOPAC method does distinguish between the two different double bonds and treats them in different environments; as the orbitals on the double bonds are different shapes and sizes in every HOMO and LUMO modelled. When the HOMO is viewed it can be seen that the double bond syn to the Cl has a large amount of electron density whereas the double bond anti to the Cl does not. The double bond syn to the Cl therefore is the most nucleophilic as it is this double bond of the two with the largest electron density.
 |
 |
 |
 |
 |
Part 2
In this section the vibrational frequencies of the C=C double bonds and C-Cl bonds were calculated to view the similarities and differences of compound 12 and the hydrogenated version of the anti double bond. The data can be viewed below and is consistent with the literature values.
| Compound 12 | Compound 12 partially hydrogenated | |
|---|---|---|
| Bond | Frequency / cm-1 | Frequency / cm-1 |
| C-Cl | 770.084 | 774.957 |
| Syn C=C | 1757.44 | 1758.06 |
| Anti C=C | 1737.01 |
From the calculated data it can be seen that the primary difference between the two compounds is that the partially hydrogenated compound 12 is lacking a anti C=C bond which is to be expected as there is no double bond in the partially hydrogenated compound in this location. The secondary difference is that the C-Cl bond has a slightly higher frequency in the partially hydrogenated compound than in compound 12, this is because there are no longer interactions between the Cl-C anti-bond and the C=C pi bond and so causing the C-Cl to be a stronger bond and therefore a higher frequency. The syn C=C bonds however are at similar values. In compound 12 the anti C=C bond is at a lower frequency than the syn C=C bond as there is mixing between the C-Cl anti bond and the C=C pi bond causing a lowering of the bond energy and so the frequency is decreased.[3]
 |
 |
 |
 |
 |
 |
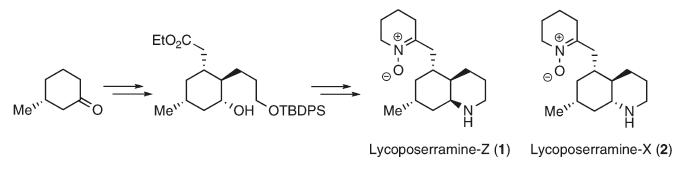
Mini Project based on Lycoposerramines-X and -Z[4]
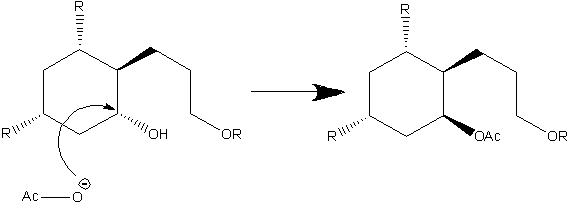
Lycoposerramines-X and -Z can be isolated from Lycopodium serratum, this can then be synthesised from (R)-3-methylcyclohexanone via Pd-catalysed Suzuki-Miyaura coupling, Johnson-Claisen rearrangement, stereoselective hydroboration - oxidation reaction and Mitsunobu reaction. The two isomers are formed due to manipulating the hydroxy group at C4, the conditions that result in the formation of the 2 isomers is shown below.
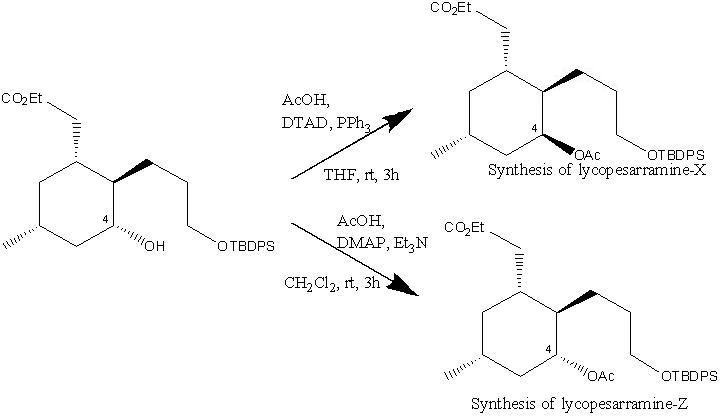
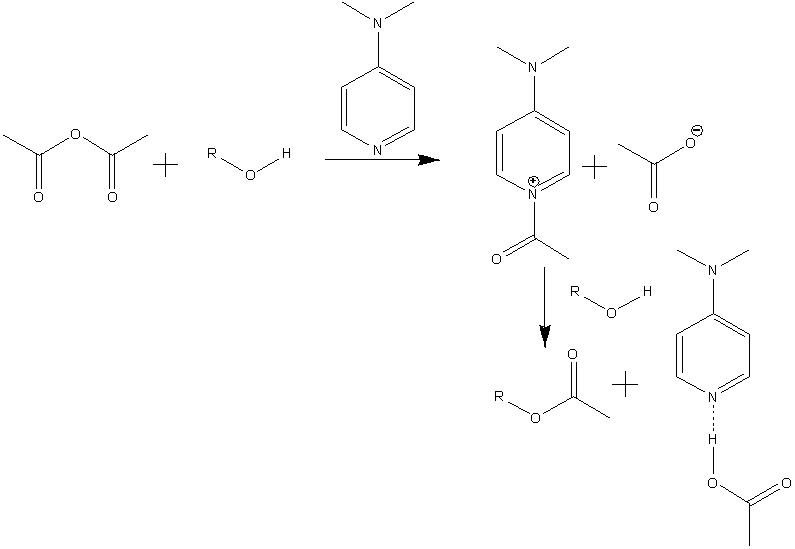
The mechanism for the formation of Lycoposerramines-Z[5] is shown below. It involves the formation of an acylpyridinium cation by reaction of the lone pair of the nitrogen from DMAP with the Ac2O, the alcohol then reacts with the acylated cataylst to form the ester product retaining the stereochemistry. This retention of stereochemistry as desired is because the alcohol attacks the acylated catalyst meaning the oxygen from the alcohol stays in the same position.
The mechanism for the formation of Lycoposerramines-X is shown below. It involves SN2 attack this therefore causes inversion to occur so the endo alcohol is converted into a exo group.
In order to differentiate between the two isomers many spectroscopic techniques can be used, the major ways are include optical rotation, circular dichroism and NMR. Optical rotation works by rotating plane polarized light one way or another depending on the isomer meaning that you can then distinguish between the two isomers, circular dichroism also works in a similar way. 1H and 13C NMR can be used and also there are multiple 2D NMR methods that can be used e.g. NOESY, HMBC and HSQC.
13C NMR Spectrum
The 13C NMR was calculated by first performing an initial geometry refinement with MM2 in ChemBio3D, the DFT=mpw1pw91 method with the 6-31G(d,p) basis set was used to optimise the geometry further and then resubmitted to perform the NMR calculation. The results from these calculations can be seen below and show the data calculated is reasonably similar to experimental values and is therefore in good agreement, with most values within 3ppm of the experimental value. Although there are two values that do disagree with the literature by large values, these being the C-13 atom which are double bonded to the N+. This therefore indicates that the molecule is in the correct conformation apart from the carbon double bonded to the nitrogen which cannot be modelled accurately.
The data for Lycoposerramine-Z (1) [6] and a NMR graph is shown below.
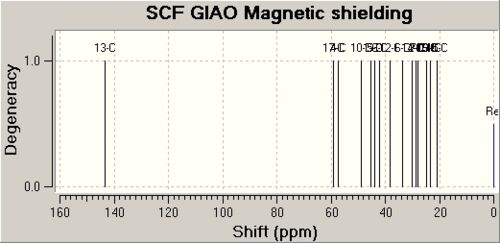
| Lycoposerramine-Z (1) | ' | ' | ' |
| Atom | Calulated Shift / ppm | Experimental Shift / ppm | Difference / ppm |
| 13 | 143.3006 | 148.7 | 5.3994 |
| 17 | 58.946 | 58.3 | 0.646 |
| 4 | 57.4231 | 56.6 | 0.8231 |
| 10 | 48.7508 | 47.9 | 0.8508 |
| 1 | 45.2014 | 41.6 | 3.6014 |
| 5 | 44.0404 | 41.4 | 2.6404 |
| 3 | 42.1037 | 40.9 | 1.2037 |
| 12 | 38.3847 | 35.8 | 2.5847 |
| 6 | 33.6363 | 29.84 | 3.7963 |
| 14 | 29.9666 | 29.81 | 0.1566 |
| 2 | 28.8062 | 26.9 | 1.9062 |
| 7 | 27.8859 | 26.7 | 1.1859 |
| 16 | 24.6559 | 23.3 | 1.3559 |
| 9 | 23.3541 | 22.7 | 0.6541 |
| 11 | 23.2358 | 21.2 | 2.0358 |
| 15 | 20.8264 | 18.9 | 1.9264 |
The data for Lycoposerramine-X (2) [7] and a NMR graph is shown below.
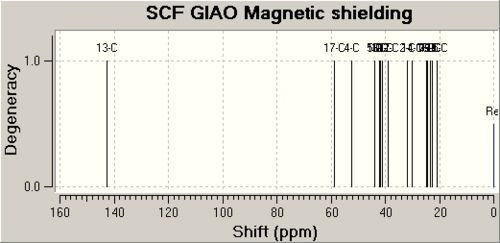
| Lycoposerramine-X (2) | ' | ' | ' |
| Atom | Calulated Shift / ppm | Experimental Shift / ppm | Difference / ppm |
| 13 | 142.6436 | 157 | 14.3564 |
| 4 | 58.9331 | 61.6 | 2.6669 |
| 17 | 52.2742 | 58.9 | 6.6258 |
| 5 | 44.0163 | 47.9 | 3.8837 |
| 10 | 43.7586 | 47.2 | 3.4414 |
| 1 | 42.1262 | 42.4 | 0.2738 |
| 3 | 41.6299 | 41.9 | 0.2701 |
| 6 | 41.095 | 39.2 | 1.895 |
| 12 | 39.0328 | 36.5 | 2.5328 |
| 2 | 32.0279 | 32 | 0.0279 |
| 14 | 29.9146 | 31.4 | 1.4854 |
| 7 | 24.7961 | 29.3 | 4.5039 |
| 9 | 24.5783 | 27.1 | 2.5217 |
| 16 | 23.2044 | 23.9 | 0.6956 |
| 11 | 22.7788 | 22.7 | 0.0788 |
| 15 | 20.9192 | 19.3 | 1.6192 |
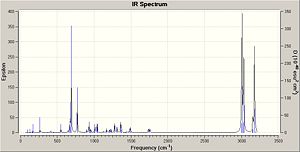
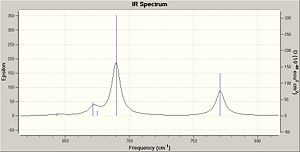
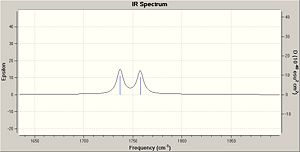
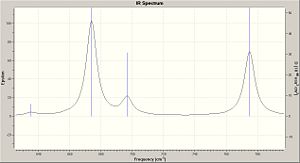
IR Spectrum
The IR spectra were calculated by first performing an initial geometry refinement with MM2 in ChemBio3D, the DFT=mpw1pw91 method with the 6-31G(d,p) basis set was used to optimise the geometry further and then resubmitted to perform the IR calculation. The IR spectra below show the usual C-H and C-C stretches and bends, however only the data where comparison could be carried out with the literature data was analysed. Therefore when the IR spectra were analysed it can be seen that the predicted values are lacking several peaks that are present in the experimental spectra. The N-H stretch is present in both spectra but at a much higher value than in the experimental spectra and one of the N=C stretches is present in both spectra at a value of 50-60 cm-1 higher than when compared with the experimental value. This shows that although the computational method can produce IR with some similar peaks it does not however predict a spectrum that can be of any use due to the lack and shift of peaks.
The data for Lycoposerramine-Z (1) and an IR spectrum is shown below.
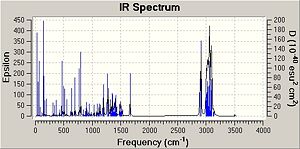
| ' | Lycoposerramine-Z (1) | ' |
| Type | Predicted Value / cm-1 | Experimental Value / cm-1 |
| N-H stretch | 3504.82 | 3275 |
| N=C stretch | N/A | 2940 |
| N=C stretch | N/A | 2914 |
| N=C stretch | 1663.77 | 1604 |
The data for Lycoposerramine-X (2) and an IR spectrum is shown below.
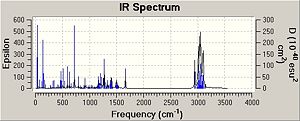
| ' | Lycoposerramine-X (2) | ' |
| Type | Predicted Value / cm-1 | Experimental Value / cm-1 |
| N-H stretch | 3521.34 | 3281 |
| N=C stretch | N/A | 3231 |
| N=C stretch | N/A | 2924 |
| N=C stretch | N/A | 2849 |
| N=C stretch | N/A | 2789 |
| N=C stretch | 1663.50 | 1619 |
Optical Rotation
Optical rotation calculations were run and submitted to SCAN however the calculation was unsuccessful, this is most likely due to the large size of the molecule causing the calculation to crash or timeout. This calculation was rerun again but again it failed. There was however not enough time remaining to attempt to fix this and rerun these calculations again.
Circular Dichrosim
Circular Dichroism calcualtions were run and submitted to SCAN however the calculation was unsuccessful, this is most likely due to the large size of the molecule causing the calculation to crash or timeout. This calculation was rerun again but again it failed. There was however not enough time remaining to attempt to fix this and rerun these calculations again.
Conclusion
This therefore shows that although the computational calculations can predict certain things it also has many shortcomings. The 13C NMR prediction as seen above was reasonably successful, however the IR spectrum which was calculated did not agree with the experimental spectrum. The optical rotation and circular dichroism calculations also failed, illustrating issues with predicting these with large molecules.
References
- ↑ 'Regio- and Stereoselective Control in the Addition of Grignard Reagents to the Pyridine Ring System' - A. G. Shultz, L. Flood and J. P. Springer, Journal of Organic Chemistry, 1986, 51, 838-841: DOI:10.1021/jo00356a016
- ↑ 'Evaluation and Prediction of the Stability of Bridgehead Olefins' - W. F. Maier and P. von Raguc Schleyer, Jounrnal of American Chemical Society, 1981, 103, 1891-1900
- ↑ 'Characteristic IR Absorption Frequencies of Organic Functional Groups' - http://www2.ups.edu/faculty/hanson/Spectroscopy/IR/IRfrequencies.html
- ↑ 'Asymmetric Total Syntheses of Cyclic Nitrone-Containing Phlegmarine-Type Lycopodium Alkaloids, Lycoposerramines-X and -Z' - T. Tanaka, N. Kogure, M. Kitajima and H. Takayama, Journal of Organic Chemistry, 2009, 74, 8675-8680
- ↑ 'The DMAP-Catalyzed Acetylation of Alcohols—A Mechanistic Study' - S. Xu, I. Held, B. Kempf, H. Mayr, W. Steglich and H. Zipse, Chemistry - A European Journal, 2005, 11, 4751-4757
- ↑ NMR Data file for Lycoposerramine-Z DOI:10042/to-2926 ]
- ↑ NMR Data file for Lycoposerramine-X DOI:10042/to-2927 ]
