Rep:Mod:FC1510
Computational Chemistry Lab: Module 2: Week 1: Part A
Optimising a BH3 Molecule
STEP 1: Optimising BH3 with a 3-21G Basis Set
The molecule BH3 was optimised by GaussView with a 3-21G basis set, the link directing to a representation. File:Bh3optimisationfc1510.log
Results: The optimised bond lengths of B-H is 1.19 Å, and the H-B-H angle is 120°.
| BH3 Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 3-21G | |
| E(RB3LYP) | -26.46226338 | a.u. |
| RMS Gradient Norm | 0.00020672 | a.u. |
| Dipole Moment | 0 | Debye |
| Point Group | D3h |
The whole calculation took 35 seconds. It has the correct point group for BH3, and the gradient is less that .001, therefore it has been correctly optimised. The final total energy of a BH3 with a 3-21G basis set is -26.46226338 atomic units, as shown on the table.
Item Table of BH3
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Observation: everything has been converged, therefore the optimisation has been completed.
STEP 2: Optimising BH3 with a 6-31G (d,p) basis set
The molecule BH3 was again optimised, this time with a different basis set, 6-31G (d,p). File:BH3OPT631G DP.LOG
Results:
The B-H bond length is 1.19 Å, and the H-B-H angle is 120°, therefore equal to the previous optimisation with the 3-21G basis set.
| BH3 Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| E(RB3LYP) | -26.61532363 | a.u. |
| RMS Gradient Norm | 0.00000235 | a.u. |
| Dipole Moment | 0 | Debye |
| Point Group | D3h |
The calculation took 19 seconds. The difference of the change of basis set is made clear by a lower final energy and a 'more flat' (closer to zero) gradient vs. the optimisation with the 3-21G basis set. The Point Group remains correct at D3h. The final total energy with these new basis set is -26.61532363 atomic units.
Item Table of BH3
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
All the items have been converged, therefore it can be said that the calculations were correctly done by the program.
Comparisons of basis sets
The first basis set reported a final total energy of -26.46226338 atomic units and the second basis set reported a final energy of -26.61532363 atomic units. The difference is 0.15306025 au. This demonstrates that different basis sets of the same molecule will lead to a significant final total energy difference. ONE MUST ALWAYS SAY WHICH BASIS SET THEY ARE USING WHEN CONDUCTING AN OPTIMISATION.
The literature value of the bond length of BH3 is 1.19 Å, and the literature structure is planar.[1]. Therefore the two different optimised molecules predicted the correct bond length and structure (therefore angle).
Optimising a TlBr3 molecule
TlBr3 was run in the Departamental SCAN Cluster. Both different atoms are large (past the third period of the periodic table), therefore the basis set used was Lan2DZ, which uses pseudo potentials (PP). File:TlBr3OPTLan2DZ.log JOB ID: 69154 Link to D-Space
Results:
The Tl-Br bond distance is 2.65 Å, and the Br-Tl-Br angle is 120°. The angle is therefore is the same as the trigonal planar system of BH3, but the bond distance is longer, which can be attributed to the much larger atoms and their orbital interaction.
| TlBr3 optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | LANL2DZ | |
| E(RB3LYP) | -91.21812851 | a.u. |
| RMS Gradient Norm | 0.0000009 | a.u. |
| Dipole Moment | 0 | Debye |
| Point Group | D3h |
The calculation took 38.2 seconds. The gradient is well below .001 au, therefore can be assumed that optimisation was reached. The final total energy of the optimisation was -91.21812851 atomic units.
Item Table
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084076D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
All the items have been converged, therefore the optimisation concluded correctly.
Bond length comparison to literature value
The optimised structure claimed that the Tl-Br bond length is 2.65 Å, but the literature value according to the CRC handbook is 2.62 Å.[1] Therefore the result is very close, but not exact.
Optimising a BBr3 molecule
BBr3 has a small atom (B) and a large one (Br). Therefore the input file had to have its basis sets specified individually. The Boron had the 6-31G(d,p) basis sets. The Bromine atoms had the Lan2DZ basis sets. File:BBr3OPTGEN.log. JOB ID:69155 Link to D-Space
Results:
The B-Br bond distance is 1.93 Å and the Br-B-Br angle is 120°. The bond distance is larger than BH3, but shorter than that of TlBr3, which makes sense since Br is larger than H, but B is smaller than Tl.
| BBr3 Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | Gen | |
| E(RB3LYP) | -64.43645296 | a.u. |
| RMS Gradient Norm | 0.00000382 | a.u. |
| Dipole Moment | 0 | Debye |
| Point Group | D3h |
The calculation took 36.6 seconds. The gradient is less than .001 au, therefore indicating that the optimisation is complete. The final total energy is -64.43645296 au.
Item Table
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027279D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Everything is converged, therefore OPTIMISATION HAS OCCURRED.
Comparing BH3, TlBr3 and BBr3
| Molecule | BH3 | BBr3 | TlBr3 |
|---|---|---|---|
| Bond Lengths calculated | 1.19 Å | 1.93 Å | 2.65 Å |
| Literature Bond Lengths [1] | 1.19 Å | 1.89 Å | 2.62 Å |
The bond lengths have been organised in order of increasing bond length to be able to analyse them more easily. There is a very similar difference in bond length between the first two and the last two. Between the first two the main change is the ligand, when the hydrogen is replaced by the bromine. With the substitution of the larger ligand, the bond distance increases. Therefore it can be concluded that the larger the ligand, the larger the bond distance. Hydrogen and bromine are similar in some ways, such as they have similar oxidation states, but they differ very largely in size, and hydrogen is in the first row, and bromine is in the fourth, therefore has a substantial larger orbital and quantity of electrons.
On the other hand the main difference between the 2nd and 3rd molecules is that the central element is changed, from B to Tl. They are both in the same group, and therefore share some similar qualities, but Tl is four rows below B, and is therefore much larger. The addition of the larger central element leads to the longer bond, therefore the relationship is that the larger the central element, the longer the bond.
Both of the points above explain why the longest bond between all three different molecules is the one with the largest central element between all of them (Tl) and and the largest ligand (Br).
Computational Chemistry Lab: Module 2: Week 1: Part B
Frequency Analysis of BH3
A Frequency analysis was run on the 6-31G (d,p) optimised BH3 structure. File:FC1510 BH3 FREQ.LOG
Results:
| BH3 Frequency | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| E(RB3LYP) | -26.61532363 | a.u. |
| RMS Gradient Norm | 0.00000237 | a.u. |
| Dipole Moment | 0 | Debye |
| Point Group | D3h |
The calculation took 22 seconds. The final total energy is the same as the original BH3 optimised structure: this is correct as the frequency analysis should not have changed that.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.323374D-10
Optimization completed.
-- Stationary point found.
Everything is converged: proof that the calculation happened correctly.
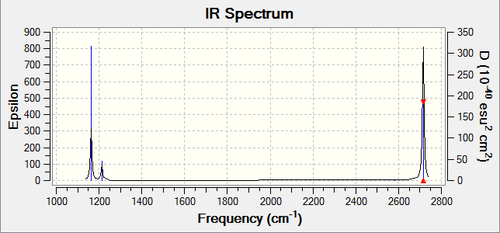
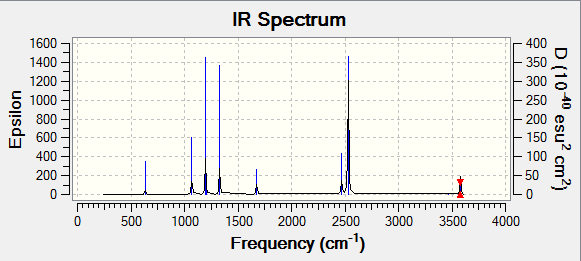
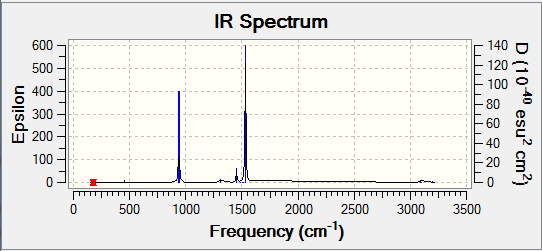
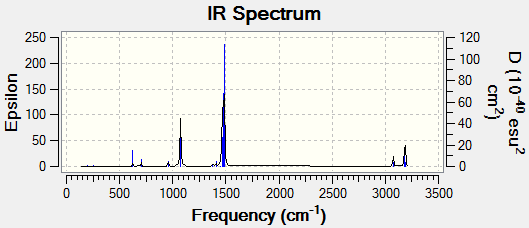
Frequency Analysis:
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824 Low frequencies --- 1163.0003 1213.1853 1213.1880
Frequency Vibrations
IR Spectra
The spectra was drawn by the Gaussian Program:
From the table we know that there are six vibrations, but the graph only seems to show three. This is because three of the vibrations have large intensities (vibrations no. 1, 5 and 6) compared the other three (vibrations no. 2, 3 and 4).
Frequency Analysis of TlBr3
Ran a frequency analysis of the PP optimised structure of TlBr3. File:Fc1510 TlBr3 freq.log. JOB ID:69189 Link to D-Space
Results:
| TlBr3 frequency analysis | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | LANL2DZ | |
| E(RB3LYP) | -91.21812851 | a.u. |
| RMS Gradient Norm | 0.00000088 | a.u. |
| Dipole Moment | 0 | Debye |
| Point Group | D3h |
The calculation took 20.9 seconds. The final total energy remains the same as before the frequency analysis, which is at it should be.
Item Table
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
-- Stationary point found.
Everything is converged.
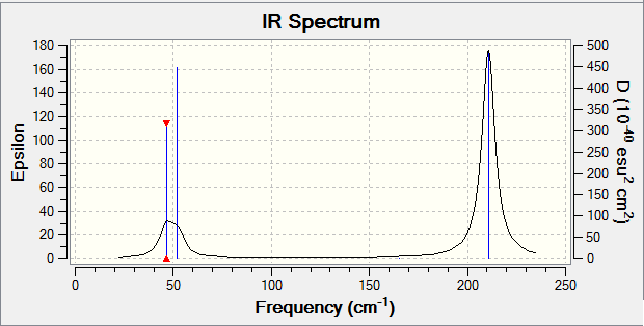
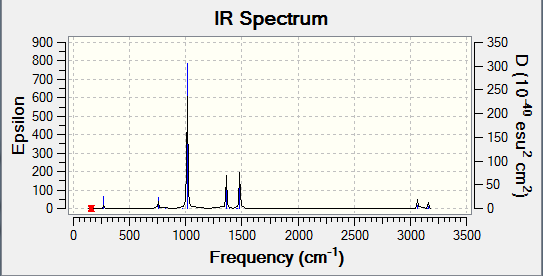
Frequency Analysis
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
There is at least a 15 cm -1 difference between the low frequencies: Correct and therefore representative that the optimised molecule has the correct set of minima requirements.
IR Spectra
Although there are 6 vibrations, again a reduced amount are shown (only two). When looking at the intensities, there are only two that are relatively larger that the others, explaining the spectra.
Computational Chemistry Lab: Module 2: Week 1: Part C
MO of BH3
BH3 was run through the Departmental SCAN Cluster. File:BH3MO.log. File:BH3MO.fchk JOB ID: 69198 Link to D-Space
Produced the MO diagrams using Gaussian and compared the MO's with LCAO theory:
Apart from the mode of representing the orbitals, this diagram shows that the MO and LCAO match-up. Therefore it is also a solid demonstration that MO Theory works as an accurate model.
Computational Lab Module 2: Week 1: Part D
NH3 Analysis
Optimising NH3
Ran an optimisation of NH3 with Gaussian. File:NH3OPT631G DP.LOG
Results: The N-H bond length is 1.02 Å and the H-N-H angle is 105.7°. The literature value[1] of the N-H bond is 1.01 Å and the H-N-H bond is 106.7°.
| NH3 Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| E(RB3LYP) | -56.55776856 | a.u. |
| RMS Gradient Norm | 0.00000888 | a.u. |
| Dipole Moment | 1.8464 | Debye |
| Point Group | C1 |
The calculation ran for 14 seconds. The gradient is below .001 au, therefore it has been properly optimised. The final total energy is -56.55776856 atomic units. The optimisation claims that the point group is C1, which is incorrect, but that is a common mistake for Gaussian.
Item Table
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000088 0.001800 YES
RMS Displacement 0.000056 0.001200 YES
Predicted change in Energy=-1.776783D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7413 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7486 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7479 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8631 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
All the forces have been converged, therefore the optimisation calculations are 'complete'.
Frequency Analysis of NH3
Performed a frequency analysis on Gaussian of the NH3 molecule. File:FC1510 NH3 FREQ.LOG
Results:
| NH3 Frequency | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| E(RB3LYP) | -56.55776856 | a.u. |
| RMS Gradient Norm | 0.00000891 | a.u. |
| Dipole Moment | 1.8464 | Debye |
| Point Group | C1 |
Calculation took 27 seconds. The final total energy remained the same.
Item Table
Item Value Threshold Converged?
Maximum Force 0.000021 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000077 0.001800 YES
RMS Displacement 0.000039 0.001200 YES
Predicted change in Energy=-1.610513D-09
Optimization completed.
-- Stationary point found.
Everything is converged, as always proving that the optimisation is complete.
Frequency Analysis:
Low frequencies:
Low frequencies --- -30.6683 -0.0008 0.0005 0.0010 20.3158 28.3272 Low frequencies --- 1089.5567 1694.1245 1694.1869
There are no negative frequencies, and more than a 15 cm-1 difference, therefore these are allowed frequencies.
IR Spectra
MO's of NH3
MOs of NH3 were analysed. File:NH3MO.log. File:NH3MO.fchk. Link to D-Space
NBO Analysis of NH3
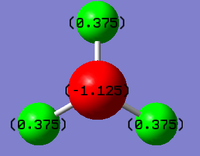
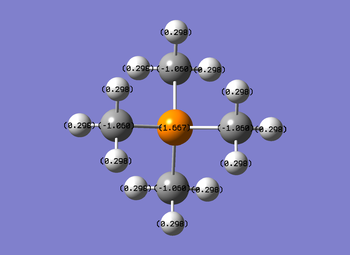
Picture of NH3 representing the charges of the atoms. Bright green represents highly positive (1.125) and bright red a highly negative charge (-1.125).
Therefore this NBO of NH3 shows that the H atoms have a relatively positive charge and the N atom has a strong negative charge.
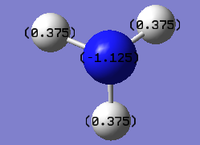
By changing the settings in Gaussian, one can actually see the specific charge of the molecule:
Therefore can see that the negative charge is concentrated in the N atom, and the positive charge is distributed in thirds throughout the three H atoms.
Computational Chemistry Lab: Module 2: Week 1: Part E
Association Energy of NH3BH3
Optimisation
NH3BH3 was optimised using Gaussian. File:NH3BH3OPT.LOG
Results:
The B-H bond length is 1.21 Å, with a H-B-H angle of 108°. The N-H bond distance in turn is 1.02 Å with an angle of 113.9°. This difference of bond length and angle makes sense since nitrogen is more electronegative than boron. Finally the bond length of N-B is 1.67 Å.
In the literature, CRC Handbook[1], the B-H bond length is 1.26 Å, the N-H bond is 1.05 Å and the N-B bond length is 1.43 Å. Therefore the results are not ideal compared to the literature.
| NH3BH3 Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| E(RB3LYP) | -83.22469032 | a.u. |
| RMS Gradient Norm | 0.00005935 | a.u. |
| Dipole Moment | 5.5648 | Debye |
| Point Group | C1 |
The calculation took 2 minutes and 8 seconds. The gradient is below .001 atomic units, marking that the optimisation happened. The final total energy is -83.22469032 atomic units.
Item Table
Item Value Threshold Converged?
Maximum Force 0.000121 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000508 0.001800 YES
RMS Displacement 0.000294 0.001200 YES
Predicted change in Energy=-1.611643D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.0186 -DE/DX = -0.0001 !
! R2 R(2,8) 1.0186 -DE/DX = -0.0001 !
! R3 R(3,8) 1.0186 -DE/DX = -0.0001 !
! R4 R(4,7) 1.21 -DE/DX = -0.0001 !
! R5 R(5,7) 1.21 -DE/DX = -0.0001 !
! R6 R(6,7) 1.21 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6681 -DE/DX = -0.0001 !
! A1 A(4,7,5) 113.8796 -DE/DX = 0.0 !
! A2 A(4,7,6) 113.8796 -DE/DX = 0.0 !
! A3 A(4,7,8) 104.5972 -DE/DX = 0.0 !
! A4 A(5,7,6) 113.8728 -DE/DX = 0.0 !
! A5 A(5,7,8) 104.591 -DE/DX = 0.0 !
! A6 A(6,7,8) 104.591 -DE/DX = 0.0 !
! A7 A(1,8,2) 107.873 -DE/DX = 0.0 !
! A8 A(1,8,3) 107.873 -DE/DX = 0.0 !
! A9 A(1,8,7) 111.0212 -DE/DX = 0.0 !
! A10 A(2,8,3) 107.8692 -DE/DX = 0.0 !
! A11 A(2,8,7) 111.0298 -DE/DX = 0.0 !
! A12 A(3,8,7) 111.0297 -DE/DX = 0.0 !
! D1 D(4,7,8,1) 179.9998 -DE/DX = 0.0 !
! D2 D(4,7,8,2) -60.0005 -DE/DX = 0.0 !
! D3 D(4,7,8,3) 60.0001 -DE/DX = 0.0 !
! D4 D(5,7,8,1) -59.9967 -DE/DX = 0.0 !
! D5 D(5,7,8,2) 60.003 -DE/DX = 0.0 !
! D6 D(5,7,8,3) -179.9964 -DE/DX = 0.0 !
! D7 D(6,7,8,1) 59.9963 -DE/DX = 0.0 !
! D8 D(6,7,8,2) 179.996 -DE/DX = 0.0 !
! D9 D(6,7,8,3) -60.0034 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Everything is converged, therefore the optimisation happened correctly.
Frequency Analysis
Performed a frequency analysis via Gauassian on the NH3BH3. File:FC1510 NH3BH3 FREQ.LOG.
Results:
| NH3BH3 Frequency | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| E(RB3LYP) | -83.22469032 | a.u. |
| RMS Gradient Norm | 0.00005932 | a.u. |
| Dipole Moment | 5.5648 | Debye |
| Point Group | C1 |
The gradient is still small and the final total energy remains the same.
Item Table
Item Value Threshold Converged?
Maximum Force 0.000113 0.000450 YES
RMS Force 0.000059 0.000300 YES
Maximum Displacement 0.000583 0.001800 YES
RMS Displacement 0.000343 0.001200 YES
Predicted change in Energy=-1.713423D-07
Optimization completed.
-- Stationary point found.
Frequencies
Low frequencies --- -0.0015 -0.0012 -0.0009 18.5275 23.7762 41.0207 Low frequencies --- 266.2847 632.2308 639.8257
Small values for the first set of low frequencies, and no negative values for the second batch of low frequencies: Correct!
IR Spectra
Following the 3N-6 rule, NH3BH3 has 18 different vibrations.
Comparisons of Energies
E(NH3)= -56.55776856 au
E(BH3)= -26.61532363 au
E(NH3BH3)= -83.22469032
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]
ΔE=(-83.22469032 au)-[(-56.55776856 au)+(-26.61532363 au)]
ΔE= -0.05159813 au
Therefore in kJ/Mol=-135 kJ/mol. This is also the dissociation energy.
Computational Lab: Module 2: Week 2: Ionic Liquids: Designer Solvents, Part 1: Comparison of selected "onium" cations
[N(CH3)4]+
Optimisation
[N(CH3)4}+ was optimised using the 6-31G (d,p) basis set with the HPC services. File:NCH34 OPT2.log.
Job: 69568. Link to d-space
Results:
Once with the optimised molecule could find out that:
Bond Lengths: N-C bond: 1.51 Å, C-H bond: 1.09 Å
Bond Angles: C-N-C : 109.5°, H-C-H: 110.1°, and H-C-N: 108.9°.
Comment on geometries:
Since all C-N-C bond angles are 109.5° after optimisations demonstrates that N(CH3)4+ has a tetrahedral structure at the nitrogen centre, therefore will have sp3 hybridization at the same point. The bond length make sense when considering valence electrons and orbital size. Carbon and hydrogen both have smaller orbitals than nitrogen, therefore will have better orbital overlap between themselves than carbon and nitrogen, therefore explaining why they have a shorter bond between themselves.
| N(CH3)4+ Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -214.1812726 | a.u. |
| RMS Gradient Norm | 0.000021 | a.u. |
| Dipole Moment | 1.9491 | Debye |
| Point Group | C1 |
The calculation took 5 minutes, 45.7 seconds. The gradient is less than .001 au, which is correct for optimisations. The final total energy was -214.1812726 au.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000074 0.000450 YES
RMS Force 0.000017 0.000300 YES
Maximum Displacement 0.000673 0.001800 YES
RMS Displacement 0.000176 0.001200 YES
Predicted change in Energy=-4.690832D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.0902 -DE/DX = 0.0 !
! R2 R(1,3) 1.0902 -DE/DX = 0.0 !
! R3 R(1,4) 1.0902 -DE/DX = 0.0 !
! R4 R(1,17) 1.5095 -DE/DX = -0.0001 !
! R5 R(5,6) 1.0902 -DE/DX = 0.0 !
! R6 R(5,7) 1.0902 -DE/DX = 0.0 !
! R7 R(5,8) 1.0902 -DE/DX = 0.0 !
! R8 R(5,17) 1.5095 -DE/DX = -0.0001 !
! R9 R(9,10) 1.0902 -DE/DX = 0.0 !
! R10 R(9,11) 1.0902 -DE/DX = 0.0 !
! R11 R(9,12) 1.0902 -DE/DX = 0.0 !
! R12 R(9,17) 1.5095 -DE/DX = -0.0001 !
! R13 R(13,14) 1.0902 -DE/DX = 0.0 !
! R14 R(13,15) 1.0902 -DE/DX = 0.0 !
! R15 R(13,16) 1.0902 -DE/DX = 0.0 !
! R16 R(13,17) 1.5096 -DE/DX = -0.0001 !
! A1 A(2,1,3) 110.0477 -DE/DX = 0.0 !
! A2 A(2,1,4) 110.0523 -DE/DX = 0.0 !
! A3 A(2,1,17) 108.9032 -DE/DX = 0.0 !
! A4 A(3,1,4) 110.0524 -DE/DX = 0.0 !
! A5 A(3,1,17) 108.8703 -DE/DX = 0.0 !
! A6 A(4,1,17) 108.8822 -DE/DX = 0.0 !
! A7 A(6,5,7) 110.0524 -DE/DX = 0.0 !
! A8 A(6,5,8) 110.0478 -DE/DX = 0.0 !
! A9 A(6,5,17) 108.9032 -DE/DX = 0.0 !
! A10 A(7,5,8) 110.0524 -DE/DX = 0.0 !
! A11 A(7,5,17) 108.8822 -DE/DX = 0.0 !
! A12 A(8,5,17) 108.8702 -DE/DX = 0.0 !
! A13 A(10,9,11) 110.0621 -DE/DX = 0.0 !
! A14 A(10,9,12) 110.075 -DE/DX = 0.0 !
! A15 A(10,9,17) 108.8709 -DE/DX = 0.0 !
! A16 A(11,9,12) 110.0748 -DE/DX = 0.0 !
! A17 A(11,9,17) 108.8709 -DE/DX = 0.0 !
! A18 A(12,9,17) 108.8529 -DE/DX = 0.0 !
! A19 A(14,13,15) 110.0291 -DE/DX = 0.0 !
! A20 A(14,13,16) 110.0571 -DE/DX = 0.0 !
! A21 A(14,13,17) 108.8959 -DE/DX = 0.0 !
! A22 A(15,13,16) 110.057 -DE/DX = 0.0 !
! A23 A(15,13,17) 108.8959 -DE/DX = 0.0 !
! A24 A(16,13,17) 108.8733 -DE/DX = 0.0 !
! A25 A(1,17,5) 109.4776 -DE/DX = 0.0 !
! A26 A(1,17,9) 109.4528 -DE/DX = 0.0 !
! A27 A(1,17,13) 109.4814 -DE/DX = 0.0 !
! A28 A(5,17,9) 109.4527 -DE/DX = 0.0 !
! A29 A(5,17,13) 109.4814 -DE/DX = 0.0 !
! A30 A(9,17,13) 109.4813 -DE/DX = 0.0 !
! D1 D(2,1,17,5) -60.0378 -DE/DX = 0.0 !
! D2 D(2,1,17,9) 179.9923 -DE/DX = 0.0 !
! D3 D(2,1,17,13) 59.9849 -DE/DX = 0.0 !
! D4 D(3,1,17,5) 59.9603 -DE/DX = 0.0 !
! D5 D(3,1,17,9) -60.0097 -DE/DX = 0.0 !
! D6 D(3,1,17,13) 179.983 -DE/DX = 0.0 !
! D7 D(4,1,17,5) 179.9515 -DE/DX = 0.0 !
! D8 D(4,1,17,9) 59.9816 -DE/DX = 0.0 !
! D9 D(4,1,17,13) -60.0258 -DE/DX = 0.0 !
! D10 D(6,5,17,1) 60.0186 -DE/DX = 0.0 !
! D11 D(6,5,17,9) -180.0114 -DE/DX = 0.0 !
! D12 D(6,5,17,13) -60.0041 -DE/DX = 0.0 !
! D13 D(7,5,17,1) -179.9706 -DE/DX = 0.0 !
! D14 D(7,5,17,9) -60.0006 -DE/DX = 0.0 !
! D15 D(7,5,17,13) 60.0067 -DE/DX = 0.0 !
! D16 D(8,5,17,1) -59.9794 -DE/DX = 0.0 !
! D17 D(8,5,17,9) 59.9906 -DE/DX = 0.0 !
! D18 D(8,5,17,13) -180.0021 -DE/DX = 0.0 !
! D19 D(10,9,17,1) 179.9962 -DE/DX = 0.0 !
! D20 D(10,9,17,5) 60.0109 -DE/DX = 0.0 !
! D21 D(10,9,17,13) -59.9964 -DE/DX = 0.0 !
! D22 D(11,9,17,1) -60.007 -DE/DX = 0.0 !
! D23 D(11,9,17,5) 180.0077 -DE/DX = 0.0 !
! D24 D(11,9,17,13) 60.0004 -DE/DX = 0.0 !
! D25 D(12,9,17,1) 59.9945 -DE/DX = 0.0 !
! D26 D(12,9,17,5) -59.9907 -DE/DX = 0.0 !
! D27 D(12,9,17,13) 180.0019 -DE/DX = 0.0 !
! D28 D(14,13,17,1) 59.9988 -DE/DX = 0.0 !
! D29 D(14,13,17,5) -179.9808 -DE/DX = 0.0 !
! D30 D(14,13,17,9) -59.9911 -DE/DX = 0.0 !
! D31 D(15,13,17,1) 179.9851 -DE/DX = 0.0 !
! D32 D(15,13,17,5) -59.9946 -DE/DX = 0.0 !
! D33 D(15,13,17,9) 59.9952 -DE/DX = 0.0 !
! D34 D(16,13,17,1) -60.0081 -DE/DX = 0.0 !
! D35 D(16,13,17,5) 60.0122 -DE/DX = 0.0 !
! D36 D(16,13,17,9) 180.002 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
All the forces are converged, corroborating that the optimisation finalized.
Frequency Analysis
Performed the frequency analysis using the HCP database. File:NCH34 frequency.log.
Job ID:69587 Link to D-Space
Results:
| N(CH3)4+ Frequency | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -214.1812726 | a.u. |
| RMS Gradient Norm | 0.00002102 | a.u. |
| Dipole Moment | 1.9491 | Debye |
| Point Group | C1 |
Calculations took 10 minutes, 7.1 seconds. The final total energy remains the same as the opimisation and the gradient is below .001 au, therefore it occured correctly according to the summary table.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000073 0.000450 YES
RMS Force 0.000021 0.000300 YES
Maximum Displacement 0.000393 0.001800 YES
RMS Displacement 0.000153 0.001200 YES
Predicted change in Energy=-4.555770D-08
Optimization completed.
-- Stationary point found.
Everything is converged.
Frequency Analysis: From the results found that the frequencies measurements are:
Low frequencies --- -13.0274 0.0009 0.0009 0.0010 6.1464 11.9065 Low frequencies --- 179.8748 278.8589 285.6936
The first line of frequencies are +/- 15 cm-1, as it should be, and the lower set of frequencies are also allowed as they are not negative. These results demonstrate that the optimisation has minima values.
IR Spectra:
MO analysis
Proceeded to do the MO calculation in the HCP database, Link to D-Space.
The five MO's for N(CH3)4+, ranging from strongly bonding to strongly anti-bonding
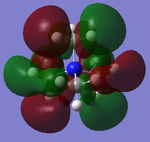
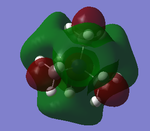
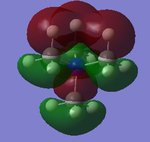
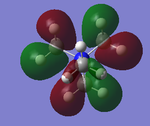
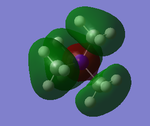
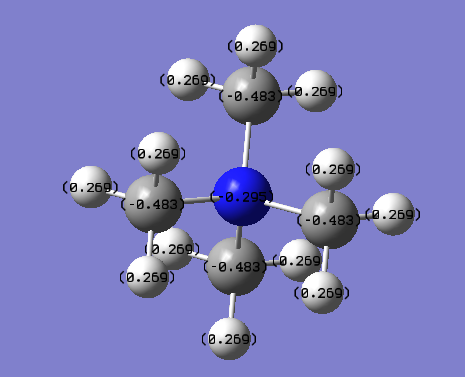
NBO Analysis
Using the .log file of the same energy run as for the MO analysis (therefore the same D-space link).
Qualitative view of [N(CH3)4+, using the same range as before (bright green for 1.125 and bright red for -1.125)
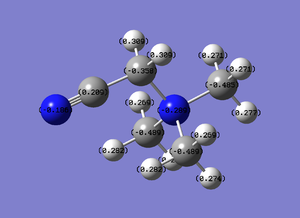
Therefore from the representation can see that all the hydrogens seem to be a dark green (therefore slightly positive charge) and nitrogen and carbon dark green (slightly negative charge). It would seem that the nitrogen is a slightly 'darker' red, therefore less negatively charged, but to find out more one would have to look at the quantitative view:
The file diagram shows that the charge on the hydrogens are all .269 (positive), the carbons all have a charge of (-.483) and the nitrogen, as noticed above in the qualitative diagram, is a little bit less negative at (-.295). This diagrams insinuates that the cation's positive charge is carried by the hydrogens on the outside, as all the other atom types have a negative charge.
Analyzing the bond contribution of the C-N analysis:
The relevant bond contribution from the NBO analysis:
4. (1.98453) BD ( 1) C 1 - N 17
( 33.65%) 0.5801* C 1 s( 20.77%)p 3.81( 79.06%)d 0.01( 0.16%)
0.0003 0.4551 -0.0237 0.0026 0.3016
0.0128 -0.7262 -0.0308 0.4135 0.0175
-0.0195 0.0111 -0.0268 -0.0194 -0.0071
( 66.35%) 0.8146* N 17 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 0.5000 -0.0007 0.0000 -0.2937
0.0001 0.7077 -0.0001 -0.4032 0.0000
-0.0086 0.0049 -0.0117 -0.0085 -0.0031
What that excerpt is trying to say, is that in the C-N bond, 33.65% is contributed by the carbon (which has a hybridisation of 20.77%s+79.06%p+.16%d) and 66.35% from the nitrogen (hybridisation 25%s+75%p).
Since the nitrogen contributes more to the bond than carbon, it explains why the nitrogen has 'less of a negative charge' than the carbon (-.265 (N) vs. -.483).
Comparison to the 'formal' picture
The formal picture of [NR4]+ shows the positive charge of the molecule on the nitrogen. By the observations just done for the NBO we know this is not true. It was discovered that the nitrogen does not have a positive charge, but on the contrary it has a slightly negative one. In reality what happens is that the hydrogens carry the positive charge instead, distributed equally among all of them.
P(CH3)4+
Optimisation
P(CH3)4+ was optimised using the HCP database. File:PCH34 OPT4.log
JOB ID: 70310. Link to D-Space
Results:
Bond Lengths: P-C: 1.82 Å, C_H: 1.09 Å
Bond Angles: C-P-C: There is a variation of angles, but all approaximately just above or below 109.5°, H-C-H: all approximately 109.0°.
Summary of Results:
| [P(CH3)4]+ Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -500.8270117 | a.u. |
| RMS Gradient Norm | 0.00000079 | a.u. |
| Dipole Moment | 4.6185 | Debye |
| Point Group | C1 |
The calculation took 18 minutes, 38.4 seconds. The final total energy is -500.82701172 au. The gradient is well below .001, therefore the molecule was optimised.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000001 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000036 0.000060 YES
RMS Displacement 0.000011 0.000040 YES
Predicted change in Energy=-1.360699D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.0933 -DE/DX = 0.0 !
! R2 R(1,3) 1.0933 -DE/DX = 0.0 !
! R3 R(1,4) 1.0933 -DE/DX = 0.0 !
! R4 R(1,17) 1.8164 -DE/DX = 0.0 !
! R5 R(5,6) 1.0933 -DE/DX = 0.0 !
! R6 R(5,7) 1.0933 -DE/DX = 0.0 !
! R7 R(5,8) 1.0933 -DE/DX = 0.0 !
! R8 R(5,17) 1.8164 -DE/DX = 0.0 !
! R9 R(9,10) 1.0933 -DE/DX = 0.0 !
! R10 R(9,11) 1.0933 -DE/DX = 0.0 !
! R11 R(9,12) 1.0933 -DE/DX = 0.0 !
! R12 R(9,17) 1.8164 -DE/DX = 0.0 !
! R13 R(13,14) 1.0933 -DE/DX = 0.0 !
! R14 R(13,15) 1.0933 -DE/DX = 0.0 !
! R15 R(13,16) 1.0933 -DE/DX = 0.0 !
! R16 R(13,17) 1.8164 -DE/DX = 0.0 !
! A1 A(2,1,3) 109.0105 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.0116 -DE/DX = 0.0 !
! A3 A(2,1,17) 109.9307 -DE/DX = 0.0 !
! A4 A(3,1,4) 109.0093 -DE/DX = 0.0 !
! A5 A(3,1,17) 109.9249 -DE/DX = 0.0 !
! A6 A(4,1,17) 109.9288 -DE/DX = 0.0 !
! A7 A(6,5,7) 109.011 -DE/DX = 0.0 !
! A8 A(6,5,8) 109.011 -DE/DX = 0.0 !
! A9 A(6,5,17) 109.924 -DE/DX = 0.0 !
! A10 A(7,5,8) 109.0104 -DE/DX = 0.0 !
! A11 A(7,5,17) 109.9298 -DE/DX = 0.0 !
! A12 A(8,5,17) 109.9298 -DE/DX = 0.0 !
! A13 A(10,9,11) 109.0115 -DE/DX = 0.0 !
! A14 A(10,9,12) 109.0105 -DE/DX = 0.0 !
! A15 A(10,9,17) 109.9307 -DE/DX = 0.0 !
! A16 A(11,9,12) 109.0093 -DE/DX = 0.0 !
! A17 A(11,9,17) 109.9288 -DE/DX = 0.0 !
! A18 A(12,9,17) 109.9249 -DE/DX = 0.0 !
! A19 A(14,13,15) 109.0116 -DE/DX = 0.0 !
! A20 A(14,13,16) 109.0099 -DE/DX = 0.0 !
! A21 A(14,13,17) 109.9268 -DE/DX = 0.0 !
! A22 A(15,13,16) 109.0099 -DE/DX = 0.0 !
! A23 A(15,13,17) 109.9268 -DE/DX = 0.0 !
! A24 A(16,13,17) 109.9308 -DE/DX = 0.0 !
! A25 A(1,17,5) 109.4661 -DE/DX = 0.0 !
! A26 A(1,17,9) 109.4709 -DE/DX = 0.0 !
! A27 A(1,17,13) 109.4764 -DE/DX = 0.0 !
! A28 A(5,17,9) 109.4661 -DE/DX = 0.0 !
! A29 A(5,17,13) 109.4715 -DE/DX = 0.0 !
! A30 A(9,17,13) 109.4764 -DE/DX = 0.0 !
! D1 D(2,1,17,5) -179.9989 -DE/DX = 0.0 !
! D2 D(2,1,17,9) -60.0085 -DE/DX = 0.0 !
! D3 D(2,1,17,13) 60.0008 -DE/DX = 0.0 !
! D4 D(3,1,17,5) -59.9992 -DE/DX = 0.0 !
! D5 D(3,1,17,9) 59.9912 -DE/DX = 0.0 !
! D6 D(3,1,17,13) -179.9995 -DE/DX = 0.0 !
! D7 D(4,1,17,5) 59.9977 -DE/DX = 0.0 !
! D8 D(4,1,17,9) 179.9881 -DE/DX = 0.0 !
! D9 D(4,1,17,13) -60.0026 -DE/DX = 0.0 !
! D10 D(6,5,17,1) 59.9948 -DE/DX = 0.0 !
! D11 D(6,5,17,9) -59.9986 -DE/DX = 0.0 !
! D12 D(6,5,17,13) 179.9981 -DE/DX = 0.0 !
! D13 D(7,5,17,1) 179.9938 -DE/DX = 0.0 !
! D14 D(7,5,17,9) 60.0004 -DE/DX = 0.0 !
! D15 D(7,5,17,13) -60.0029 -DE/DX = 0.0 !
! D16 D(8,5,17,1) -60.0043 -DE/DX = 0.0 !
! D17 D(8,5,17,9) -179.9976 -DE/DX = 0.0 !
! D18 D(8,5,17,13) 59.9991 -DE/DX = 0.0 !
! D19 D(10,9,17,1) 59.9983 -DE/DX = 0.0 !
! D20 D(10,9,17,5) 179.9887 -DE/DX = 0.0 !
! D21 D(10,9,17,13) -60.011 -DE/DX = 0.0 !
! D22 D(11,9,17,1) -179.9983 -DE/DX = 0.0 !
! D23 D(11,9,17,5) -60.0079 -DE/DX = 0.0 !
! D24 D(11,9,17,13) 59.9924 -DE/DX = 0.0 !
! D25 D(12,9,17,1) -60.0013 -DE/DX = 0.0 !
! D26 D(12,9,17,5) 59.989 -DE/DX = 0.0 !
! D27 D(12,9,17,13) 179.9894 -DE/DX = 0.0 !
! D28 D(14,13,17,1) 59.996 -DE/DX = 0.0 !
! D29 D(14,13,17,5) -60.001 -DE/DX = 0.0 !
! D30 D(14,13,17,9) -179.9981 -DE/DX = 0.0 !
! D31 D(15,13,17,1) 179.9957 -DE/DX = 0.0 !
! D32 D(15,13,17,5) 59.9987 -DE/DX = 0.0 !
! D33 D(15,13,17,9) -59.9983 -DE/DX = 0.0 !
! D34 D(16,13,17,1) -60.0042 -DE/DX = 0.0 !
! D35 D(16,13,17,5) 179.9988 -DE/DX = 0.0 !
! D36 D(16,13,17,9) 60.0018 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Everything was converged, therefore the optimisation finalised.
Frequency Analysis
For the frequency analysis, the job type that was selected was 'Optimisation and Frequency', and was analysed with the HCP database. The change in job type was due to being able to converge the results of the frequency calculations. File:PCH34 frequency4.log.
JOB ID: 70315, Link to D-Space.
Results:
| [P(CH3)4]+ Optimisation and frequency | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -500.8270117 | a.u. |
| RMS Gradient Norm | 0.00000053 | a.u. |
| Dipole Moment | 4.6185 | Debye |
| Point Group | C1 |
The calculation took 19 minutes, 32.5 seconds. The final total energy remains the same as the original optimisation.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000000 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000045 0.000060 YES
RMS Displacement 0.000014 0.000040 YES
Predicted change in Energy=-1.140042D-11
Optimization completed.
-- Stationary point found.
Everything is converged.
Frquency Analysis
Low frequencies --- -2.5510 0.0004 0.0004 0.0024 5.1717 7.5851 Low frequencies --- 156.4493 192.0462 192.2795
The frequencies are within the +/- 15 cm-1, as required.
IR Table:
MO Analyssis
Performed MO Analysis on [P(CH3)4]+, with the HCP database. Link to D-Space.
From the calculations it was possible to use Gaussview to predict the HOMO and LUMO for [P(CH3)4]+, shown below:

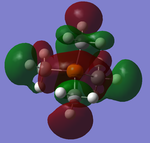
NBO Analysis
Qualitative NBO:
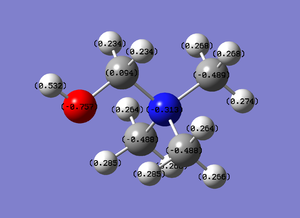
The spectrum of the NBO charge disperion is (-1.667) to (1.667). The more negative the value the brighter the red, and the more positive the value the brighter the green. Therefore in this case the Phosphorous has a very positive value.
Quantitative NBO:
To know the exact quantitative amount of the charge on the atoms, the quantitative values were calculated. As ascertained from the previous diagram, the Phosphorous has the most positive charge at 1.667, and the carbons are the only ones with negative charges, at (-1.060). The diagram calculated is shown below:
Population Analysis
Retrieved from the population analysis .log file:
4. (1.98031) BD ( 1) C 1 - P 17
( 59.57%) 0.7718* C 1 s( 25.24%)p 2.96( 74.67%)d 0.00( 0.08%)
0.0002 0.5021 0.0171 -0.0020 0.3491
-0.0064 -0.7056 0.0129 0.3560 -0.0065
-0.0166 0.0084 -0.0169 -0.0127 -0.0071
( 40.43%) 0.6358* P 17 s( 25.00%)p 2.97( 74.15%)d 0.03( 0.85%)
0.0000 0.0001 0.5000 -0.0008 0.0000
0.0000 -0.3479 0.0005 0.0000 0.7032
-0.0010 0.0000 -0.3549 0.0005 -0.0528
0.0266 -0.0538 -0.0403 -0.0226
The above information shows the results of the analysis of the C-P bond. What the calculations determined is that in this bond, 59.57% of the contribution comes from C, and 40.43% comes from P. The hybridization of the carbon contribution is (25.24%s+74.67%p), and that of the phosphorous is (25%s+75%p). To put these results into perspective, they are going to be contrasted against the other carbon-heteroatom bonds in a future section.
[S(CH3)3]+
Optimisation
Performed the optimisation in the HCP database of [S(CH3)]+, can find the file in the D-Space
Results:
Geometry observations:
Bond lengths: C-S: 1.82 Å and C-H: 1.09 Å
Bond angles: The molecule consists of three 102.7° angles of C-S, therefore they are all facing slightly 'upwards', and leave the whole of one side without any atoms.
.
The H-C-H bond angles are 109.4°.
Summary Table:
| [S(CH3)3]+ Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -517.6832736 | a.u. |
| RMS Gradient Norm | 0.00000119 | a.u. |
| Dipole Moment | 8.0651 | Debye |
| Point Group | C1 |
The calculation took 31 minutes, 2.2 seconds. The final total energy was -517.6832736 au, and with the gradient at less than .001 au, it can be assured that the molecule is has reached its optimisation energy.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000003 0.000015 YES
RMS Force 0.000001 0.000010 YES
Maximum Displacement 0.000049 0.000060 YES
RMS Displacement 0.000021 0.000040 YES
Predicted change in Energy=-1.198589D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.0916 -DE/DX = 0.0 !
! R2 R(1,3) 1.0914 -DE/DX = 0.0 !
! R3 R(1,4) 1.0916 -DE/DX = 0.0 !
! R4 R(1,13) 1.8226 -DE/DX = 0.0 !
! R5 R(5,6) 1.0916 -DE/DX = 0.0 !
! R6 R(5,7) 1.0916 -DE/DX = 0.0 !
! R7 R(5,8) 1.0914 -DE/DX = 0.0 !
! R8 R(5,13) 1.8226 -DE/DX = 0.0 !
! R9 R(9,10) 1.0916 -DE/DX = 0.0 !
! R10 R(9,11) 1.0914 -DE/DX = 0.0 !
! R11 R(9,12) 1.0916 -DE/DX = 0.0 !
! R12 R(9,13) 1.8226 -DE/DX = 0.0 !
! A1 A(2,1,3) 111.0958 -DE/DX = 0.0 !
! A2 A(2,1,4) 109.4345 -DE/DX = 0.0 !
! A3 A(2,1,13) 107.2522 -DE/DX = 0.0 !
! A4 A(3,1,4) 111.0945 -DE/DX = 0.0 !
! A5 A(3,1,13) 110.5646 -DE/DX = 0.0 !
! A6 A(4,1,13) 107.2494 -DE/DX = 0.0 !
! A7 A(6,5,7) 109.4334 -DE/DX = 0.0 !
! A8 A(6,5,8) 111.0958 -DE/DX = 0.0 !
! A9 A(6,5,13) 107.2537 -DE/DX = 0.0 !
! A10 A(7,5,8) 111.0946 -DE/DX = 0.0 !
! A11 A(7,5,13) 107.2514 -DE/DX = 0.0 !
! A12 A(8,5,13) 110.5622 -DE/DX = 0.0 !
! A13 A(10,9,11) 111.0937 -DE/DX = 0.0 !
! A14 A(10,9,12) 109.4343 -DE/DX = 0.0 !
! A15 A(10,9,13) 107.2479 -DE/DX = 0.0 !
! A16 A(11,9,12) 111.0946 -DE/DX = 0.0 !
! A17 A(11,9,13) 110.5687 -DE/DX = 0.0 !
! A18 A(12,9,13) 107.2518 -DE/DX = 0.0 !
! A19 A(1,13,5) 102.7456 -DE/DX = 0.0 !
! A20 A(1,13,9) 102.7522 -DE/DX = 0.0 !
! A21 A(5,13,9) 102.7493 -DE/DX = 0.0 !
! D1 D(2,1,13,5) -174.4826 -DE/DX = 0.0 !
! D2 D(2,1,13,9) -68.0328 -DE/DX = 0.0 !
! D3 D(3,1,13,5) -53.2144 -DE/DX = 0.0 !
! D4 D(3,1,13,9) 53.2354 -DE/DX = 0.0 !
! D5 D(4,1,13,5) 68.0505 -DE/DX = 0.0 !
! D6 D(4,1,13,9) 174.5003 -DE/DX = 0.0 !
! D7 D(6,5,13,1) 174.4979 -DE/DX = 0.0 !
! D8 D(6,5,13,9) 68.0458 -DE/DX = 0.0 !
! D9 D(7,5,13,1) -68.0346 -DE/DX = 0.0 !
! D10 D(7,5,13,9) -174.4867 -DE/DX = 0.0 !
! D11 D(8,5,13,1) 53.2301 -DE/DX = 0.0 !
! D12 D(8,5,13,9) -53.2219 -DE/DX = 0.0 !
! D13 D(10,9,13,1) -174.4768 -DE/DX = 0.0 !
! D14 D(10,9,13,5) -68.0298 -DE/DX = 0.0 !
! D15 D(11,9,13,1) -53.2115 -DE/DX = 0.0 !
! D16 D(11,9,13,5) 53.2355 -DE/DX = 0.0 !
! D17 D(12,9,13,1) 68.0575 -DE/DX = 0.0 !
! D18 D(12,9,13,5) 174.5045 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Everything got converged. Therefore [S(CH3)3]+ got optimized correctly.
Frequency Analysis
The [Optimisation + Frequency] Job type was conducted in the HCP database, with the results available in the D-Space.
Results:
Summary Table:
| [S(CH3)3]+ Optimisation and Frequency | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -517.6832736 | a.u. |
| RMS Gradient Norm | 0.0000012 | a.u. |
| Dipole Moment | 8.0651 | Debye |
| Point Group | C1 |
The calculations took 9 minutes, 20.4 seconds. The final total energy calculated is -517.6832736 atomic units, therefore the same as the optimisation. The gradient is below .001 au, therefore can be considered optimised.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000003 0.000015 YES
RMS Force 0.000001 0.000010 YES
Maximum Displacement 0.000054 0.000060 YES
RMS Displacement 0.000021 0.000040 YES
Predicted change in Energy=-1.622958D-10
Optimization completed.
-- Stationary point found.
All the items have been successfully converged, therefore the optimisation calculation can be considered finished properly.
Frequency Analysis:
From the .log file could determine the 'low frequencies':
Low frequencies --- -10.3810 -3.1183 -0.0036 -0.0031 -0.0031 5.1984 Low frequencies --- 162.2684 200.0411 200.7137
The frequencies are in the +/- 15 cm-1, and the second set are not negative, therefore they can be considered as correct guidance to the conclusion that the optimisation has found the minimas.
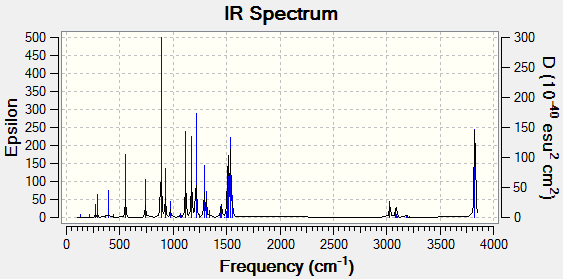
IR Table:
MO Analysis
Performed the MO analysis in the HCP database, the file with the calculation results can be found in the D-Space.
With the information gathered from the calculations, Gaussview was able to predict the MOs of the cation. Below are the MO representations of the HOMO and LUMO of [S(CH3)3]+.
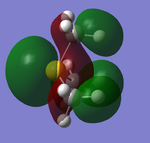

The NBO Analysis
The .log file of the MO analysis was used to calculate the NBOs of [S(CH3)3]+.
Qualitative NBO
The charge range is from -1.125 (bright red) to 1.125 (bright green).
Quantitative:
To have a representation more comparable to the other cations, one needs a quantitative view:
Population Analysis of the C-S bond
With the same .log file was able to observe the specific contributions occurring in the C-S bond:
4. (1.98631) BD ( 1) C 1 - S 13
( 48.67%) 0.6976* C 1 s( 19.71%)p 4.07( 80.16%)d 0.01( 0.14%)
0.0003 0.4437 0.0140 -0.0033 0.5677
-0.0044 0.5891 -0.0046 -0.3634 -0.0098
0.0265 -0.0167 -0.0173 -0.0010 -0.0096
( 51.33%) 0.7165* S 13 s( 16.95%)p 4.86( 82.41%)d 0.04( 0.63%)
0.0000 0.0001 0.4117 -0.0076 0.0012
0.0000 -0.5633 0.0248 0.0000 -0.5846
0.0257 0.0000 0.4038 0.0259 0.0494
-0.0430 -0.0447 -0.0018 -0.0051
Therefore the carbon contributes 48.67% of the bond population and sulfur donates 51.33% of the bond populations. The carbon hybridization is (19.71%s+80.16%p+.14%d) and the sulfur hybridization is (16.95%s+82.41%p+.63%d).
Comparing Charge Distribution
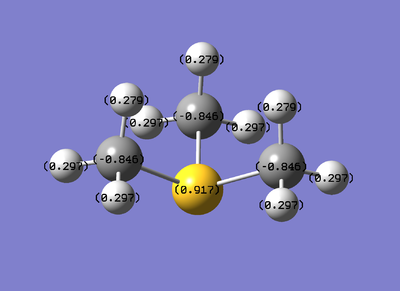
| Charge distribution diagram | Charge on central heteroatom | Charge on neighbouring carbons | Charge on hydrogens | |
|---|---|---|---|---|
| [N(CH3)4]+ | 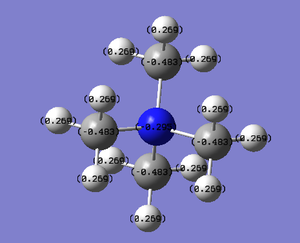
|
-0.295 | -0.483 | 0.269 |
| [P(CH3)4]+ | 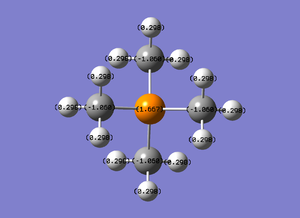
|
1.667 | -1.06 | 0.298 |
| [S(CH3)3]+ | 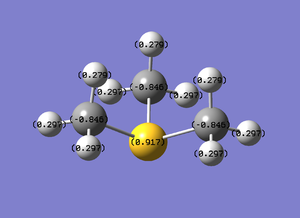
|
0.917 | -0.846 | 0.297 |
[N(CH3)4]+ is the only cation that has a negative heteroatom charge center. It seems to affect the overall charge dispersion by the fact that it has the least negative charge on the carbons compared to the other cations. Though that seems to be a trend: the more positively charged the central heteroatom, the more negatively charged the carbon is.
C-X contributions vs. charge distribution
When X=N, the contribution of the heteroatom is 66%. (The most negative charge)
When X=P, the contribution by the heteroatom is 40%. (The most positive charge)
When X=S, the contribution by the heteroatom is 51%.
Therefore it seems that the more negative the charge on the heteroatom, the more it contributes to the bond, and alternative the more positive the central element, the less it will contribute to the bond.
Computational Lab: Module 2: Week 2: Ionic Liquids: Designer Solvents, Part 2: Influence of functional groups
[N(CH3)3(CH2OH)]+
The -OH functional group was added to one of the carbons from [N(CH3)4]+, and will be run through the Gaussview program to be able to observe what changes will occur.
Optimisation
Performed the optimisation of [N(CH3)3(CH2OH)]+. Link to D-Space
Results:
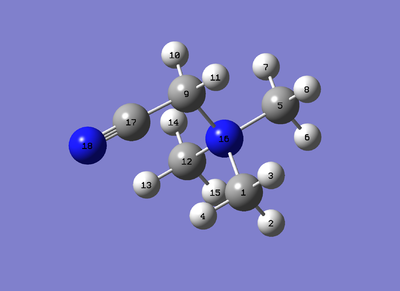
To be able to describe the varying bond lengths and angles, use the atom labelling allocated by the figure below:
(as it happens this is also a good opportunity to observe the 'optimised structure' of [N(CH3)3(CH2OH)]+)
Bond lengths:
H(18)-O(17): .96 Å. O(17)-C(9): 1.38 Å C(9)-H(10-11): 1.10 Å C(9)-N(16): 1.52 Å C(1/5/12)-N(16): 1.51 Å C(1/5/12)-H: 1.09 Å
Therefore by far the shortest bond length is that of hydroxide bond. And the longest seems to be between the carbon(9), the one bonded to the oxygenn, to the nitrogen.
The CRC Handbook[1] claims that the literature H(18)-O(17) bond length of an alcohol should be .96 Å, which is the same as the one calculated, and therefore correct.
Bond Angles:
H(18)-O(17)-C(9): 110.5° O(17)-C(9)-H(10/11): 113.7° H(10)-C(9)-H(11): 109.8° O(17)-C(9)-N(16): 106.1° C(9)-N(16)-C(1/12): 109.6° C(9)-N(16)-C(5): 108.5° C(1)-N(16)-C(12): 109.6° C(1/12)-N(16)-C(5): 109.8° For C(1/5/12), the H-C-H angles are all variant depending on which one is being measured, they range from 109.8° to 110.9°.
Summary Table:
| [N(CH3)3(CH2OH)]+ Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -289.3932191 | a.u. |
| RMS Gradient Norm | 0.00000034 | a.u. |
| Dipole Moment | 3.2806 | Debye |
| Point Group | C1 |
The calculation took 21 minutes, 41.2 seconds. The final total energy is -289.3932191 au. The calculation was successful due to the fact that the gradient's value is less than .001 au.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000001 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000011 0.000060 YES
RMS Displacement 0.000003 0.000040 YES
Predicted change in Energy=-5.835236D-12
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.0898 -DE/DX = 0.0 !
! R2 R(1,3) 1.0905 -DE/DX = 0.0 !
! R3 R(1,4) 1.0877 -DE/DX = 0.0 !
! R4 R(1,16) 1.5123 -DE/DX = 0.0 !
! R5 R(5,6) 1.0898 -DE/DX = 0.0 !
! R6 R(5,7) 1.0905 -DE/DX = 0.0 !
! R7 R(5,8) 1.0905 -DE/DX = 0.0 !
! R8 R(5,16) 1.5062 -DE/DX = 0.0 !
! R9 R(9,10) 1.0976 -DE/DX = 0.0 !
! R10 R(9,11) 1.0976 -DE/DX = 0.0 !
! R11 R(9,16) 1.5209 -DE/DX = 0.0 !
! R12 R(9,17) 1.3867 -DE/DX = 0.0 !
! R13 R(12,13) 1.0877 -DE/DX = 0.0 !
! R14 R(12,14) 1.0905 -DE/DX = 0.0 !
! R15 R(12,15) 1.0898 -DE/DX = 0.0 !
! R16 R(12,16) 1.5123 -DE/DX = 0.0 !
! R17 R(17,18) 0.9675 -DE/DX = 0.0 !
! A1 A(2,1,3) 110.0861 -DE/DX = 0.0 !
! A2 A(2,1,4) 110.8645 -DE/DX = 0.0 !
! A3 A(2,1,16) 108.3683 -DE/DX = 0.0 !
! A4 A(3,1,4) 110.2219 -DE/DX = 0.0 !
! A5 A(3,1,16) 108.6617 -DE/DX = 0.0 !
! A6 A(4,1,16) 108.5753 -DE/DX = 0.0 !
! A7 A(6,5,7) 109.8177 -DE/DX = 0.0 !
! A8 A(6,5,8) 109.8184 -DE/DX = 0.0 !
! A9 A(6,5,16) 108.5799 -DE/DX = 0.0 !
! A10 A(7,5,8) 110.244 -DE/DX = 0.0 !
! A11 A(7,5,16) 109.176 -DE/DX = 0.0 !
! A12 A(8,5,16) 109.1746 -DE/DX = 0.0 !
! A13 A(10,9,11) 109.8289 -DE/DX = 0.0 !
! A14 A(10,9,16) 106.3983 -DE/DX = 0.0 !
! A15 A(10,9,17) 113.7491 -DE/DX = 0.0 !
! A16 A(11,9,16) 106.3935 -DE/DX = 0.0 !
! A17 A(11,9,17) 113.7509 -DE/DX = 0.0 !
! A18 A(16,9,17) 106.1073 -DE/DX = 0.0 !
! A19 A(13,12,14) 110.223 -DE/DX = 0.0 !
! A20 A(13,12,15) 110.8629 -DE/DX = 0.0 !
! A21 A(13,12,16) 108.5772 -DE/DX = 0.0 !
! A22 A(14,12,15) 110.0867 -DE/DX = 0.0 !
! A23 A(14,12,16) 108.6605 -DE/DX = 0.0 !
! A24 A(15,12,16) 108.3674 -DE/DX = 0.0 !
! A25 A(1,16,5) 109.7792 -DE/DX = 0.0 !
! A26 A(1,16,9) 109.6199 -DE/DX = 0.0 !
! A27 A(1,16,12) 109.5644 -DE/DX = 0.0 !
! A28 A(5,16,9) 108.456 -DE/DX = 0.0 !
! A29 A(5,16,12) 109.7798 -DE/DX = 0.0 !
! A30 A(9,16,12) 109.6241 -DE/DX = 0.0 !
! A31 A(9,17,18) 110.4818 -DE/DX = 0.0 !
! D1 D(2,1,16,5) -61.7862 -DE/DX = 0.0 !
! D2 D(2,1,16,9) 179.1782 -DE/DX = 0.0 !
! D3 D(2,1,16,12) 58.8415 -DE/DX = 0.0 !
! D4 D(3,1,16,5) 57.8237 -DE/DX = 0.0 !
! D5 D(3,1,16,9) -61.2119 -DE/DX = 0.0 !
! D6 D(3,1,16,12) 178.4514 -DE/DX = 0.0 !
! D7 D(4,1,16,5) 177.7171 -DE/DX = 0.0 !
! D8 D(4,1,16,9) 58.6815 -DE/DX = 0.0 !
! D9 D(4,1,16,12) -61.6552 -DE/DX = 0.0 !
! D10 D(6,5,16,1) 60.2471 -DE/DX = 0.0 !
! D11 D(6,5,16,9) 179.9957 -DE/DX = 0.0 !
! D12 D(6,5,16,12) -60.2502 -DE/DX = 0.0 !
! D13 D(7,5,16,1) 179.9553 -DE/DX = 0.0 !
! D14 D(7,5,16,9) -60.2961 -DE/DX = 0.0 !
! D15 D(7,5,16,12) 59.458 -DE/DX = 0.0 !
! D16 D(8,5,16,1) -59.461 -DE/DX = 0.0 !
! D17 D(8,5,16,9) 60.2876 -DE/DX = 0.0 !
! D18 D(8,5,16,12) -179.9583 -DE/DX = 0.0 !
! D19 D(10,9,16,1) 178.3838 -DE/DX = 0.0 !
! D20 D(10,9,16,5) 58.5356 -DE/DX = 0.0 !
! D21 D(10,9,16,12) -61.3158 -DE/DX = 0.0 !
! D22 D(11,9,16,1) 61.3088 -DE/DX = 0.0 !
! D23 D(11,9,16,5) -58.5394 -DE/DX = 0.0 !
! D24 D(11,9,16,12) -178.3908 -DE/DX = 0.0 !
! D25 D(17,9,16,1) -60.1535 -DE/DX = 0.0 !
! D26 D(17,9,16,5) 179.9983 -DE/DX = 0.0 !
! D27 D(17,9,16,12) 60.1469 -DE/DX = 0.0 !
! D28 D(10,9,17,18) -63.373 -DE/DX = 0.0 !
! D29 D(11,9,17,18) 63.3893 -DE/DX = 0.0 !
! D30 D(16,9,17,18) -179.9943 -DE/DX = 0.0 !
! D31 D(13,12,16,1) 61.6327 -DE/DX = 0.0 !
! D32 D(13,12,16,5) -177.74 -DE/DX = 0.0 !
! D33 D(13,12,16,9) -58.7014 -DE/DX = 0.0 !
! D34 D(14,12,16,1) -178.4722 -DE/DX = 0.0 !
! D35 D(14,12,16,5) -57.8449 -DE/DX = 0.0 !
! D36 D(14,12,16,9) 61.1937 -DE/DX = 0.0 !
! D37 D(15,12,16,1) -58.8627 -DE/DX = 0.0 !
! D38 D(15,12,16,5) 61.7646 -DE/DX = 0.0 !
! D39 D(15,12,16,9) -179.1968 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
All the items have been convegered, therefore the optimisation has properly occurred!
Frequency Analysis
The Optimisation and Frequency analysis was undertaken in the HCP database, and the files can be located in the D-Space.
Summary Table:
| [N(CH3)3(CH2OH)]+ Optimisation and Frequency | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -289.3932191 | a.u. |
| RMS Gradient Norm | 0.00000043 | a.u. |
| Dipole Moment | 3.2806 | Debye |
| Point Group | C1 |
The calculations took 26 minutes, 6.9 seconds.
Item Table:
Item Value Threshold Converged? Maximum Force 0.000001 0.000015 YES RMS Force 0.000000 0.000010 YES Maximum Displacement 0.000021 0.000060 YES RMS Displacement 0.000005 0.000040 YES Predicted change in Energy=-9.524211D-12 Optimization completed.
Frequency analysis:
Low frequencies --- -119.2898 -4.0534 -0.0005 0.0004 0.0008 2.9261 Low frequencies --- 6.5586 129.7218 217.7265
The frequencies are not ideal.
IR Spectra:
MO Analysis
Performed the MO analysis, file in the D-Space.
Could therefore determine the HOMO and LUMO:
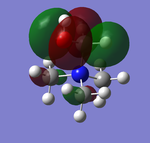
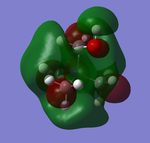
NBO Analysis
With different information from the same calculation of the MO analysis can view the charge dispersion.
Qualitative:
As before the cahrge colour range goes from -1.125 to 1.125. Therefore the oxygen seems to indicate being the one with the most negative charge.
Quantitative:
[N(CH3)3(CH2CN)]+
Another different functional group was added to [N(CH3)4]+, this time -CN. The same calculations will be run through that were used for [N(CH3)3(CH2OH)]+, and comparison will be drawn.
Optimisation
Optimised [N(CH3)3(CH2CN)]+, can find the .log file in the D-Space
Results:
To be able to describe the bond lengths and angles of [N(CH3)3(CH2CN)]+, there needs to be a numbering system on the optimised structure:
Bond lengths:
N(18)-C(17): 1.16 Å C(17)-C(9): 1.45 Å C(9)-H(10/11): 1.09 Å C(9)-N(16): 1.53 Å N(16)-C(1/5/12): 1.51 Å C(1/5/12)-H: 1.09 Å
According to the CRC Handbook [1], the N(18)-C(17) bond length should be 1.16 Å, therefore the optimised bond length is equal to the literature bond length.
Bond Angles:
N(18)-C(17)-C(9): 178.9° C(17)-C(9)-H(10/11): 110.4° C(17)-C(9)-N(16): 111.7° C(9)-N(16)-C(1/12): 110.1° C(9)-N(16)-C(5): 108.0° The H-C(1/5/12)-H are quite variant in this molecule as well.
Summary Table:
| [N(CH3)3(CH2CN)]+ Optimisation | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -306.3937605 | a.u. |
| RMS Gradient Norm | 0.00000016 | a.u. |
| Dipole Moment | 2.0625 | Debye |
| Point Group | C1 |
The calculations took 26 minutes, 54.1 seconds. The final total energy is -306.3937605 au, and the molecule got fully optimised due to the gradient being smaller than .001 au.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000000 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000003 0.000060 YES
RMS Displacement 0.000001 0.000040 YES
Predicted change in Energy=-6.199157D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.0897 -DE/DX = 0.0 !
! R2 R(1,3) 1.0905 -DE/DX = 0.0 !
! R3 R(1,4) 1.0897 -DE/DX = 0.0 !
! R4 R(1,16) 1.5139 -DE/DX = 0.0 !
! R5 R(5,6) 1.0897 -DE/DX = 0.0 !
! R6 R(5,7) 1.0904 -DE/DX = 0.0 !
! R7 R(5,8) 1.0904 -DE/DX = 0.0 !
! R8 R(5,16) 1.5116 -DE/DX = 0.0 !
! R9 R(9,10) 1.0935 -DE/DX = 0.0 !
! R10 R(9,11) 1.0935 -DE/DX = 0.0 !
! R11 R(9,16) 1.5264 -DE/DX = 0.0 !
! R12 R(9,17) 1.4599 -DE/DX = 0.0 !
! R13 R(12,13) 1.0897 -DE/DX = 0.0 !
! R14 R(12,14) 1.0905 -DE/DX = 0.0 !
! R15 R(12,15) 1.0897 -DE/DX = 0.0 !
! R16 R(12,16) 1.5139 -DE/DX = 0.0 !
! R17 R(17,18) 1.1596 -DE/DX = 0.0 !
! A1 A(2,1,3) 110.0988 -DE/DX = 0.0 !
! A2 A(2,1,4) 110.2434 -DE/DX = 0.0 !
! A3 A(2,1,16) 108.2881 -DE/DX = 0.0 !
! A4 A(3,1,4) 110.2836 -DE/DX = 0.0 !
! A5 A(3,1,16) 108.8503 -DE/DX = 0.0 !
! A6 A(4,1,16) 109.03 -DE/DX = 0.0 !
! A7 A(6,5,7) 109.9425 -DE/DX = 0.0 !
! A8 A(6,5,8) 109.944 -DE/DX = 0.0 !
! A9 A(6,5,16) 108.4565 -DE/DX = 0.0 !
! A10 A(7,5,8) 110.3236 -DE/DX = 0.0 !
! A11 A(7,5,16) 109.069 -DE/DX = 0.0 !
! A12 A(8,5,16) 109.0682 -DE/DX = 0.0 !
! A13 A(10,9,11) 108.7308 -DE/DX = 0.0 !
! A14 A(10,9,16) 107.7745 -DE/DX = 0.0 !
! A15 A(10,9,17) 110.3656 -DE/DX = 0.0 !
! A16 A(11,9,16) 107.7723 -DE/DX = 0.0 !
! A17 A(11,9,17) 110.3666 -DE/DX = 0.0 !
! A18 A(16,9,17) 111.7238 -DE/DX = 0.0 !
! A19 A(13,12,14) 110.283 -DE/DX = 0.0 !
! A20 A(13,12,15) 110.2448 -DE/DX = 0.0 !
! A21 A(13,12,16) 109.0263 -DE/DX = 0.0 !
! A22 A(14,12,15) 110.0995 -DE/DX = 0.0 !
! A23 A(14,12,16) 108.8519 -DE/DX = 0.0 !
! A24 A(15,12,16) 108.2888 -DE/DX = 0.0 !
! A25 A(1,16,5) 109.4973 -DE/DX = 0.0 !
! A26 A(1,16,9) 110.1004 -DE/DX = 0.0 !
! A27 A(1,16,12) 109.6494 -DE/DX = 0.0 !
! A28 A(5,16,9) 107.9664 -DE/DX = 0.0 !
! A29 A(5,16,12) 109.4998 -DE/DX = 0.0 !
! A30 A(9,16,12) 110.1015 -DE/DX = 0.0 !
! A31 L(9,17,18,4,-1) 178.9951 -DE/DX = 0.0 !
! A32 L(9,17,18,4,-2) 180.5658 -DE/DX = 0.0 !
! D1 D(2,1,16,5) -60.4565 -DE/DX = 0.0 !
! D2 D(2,1,16,9) -179.0126 -DE/DX = 0.0 !
! D3 D(2,1,16,12) 59.7041 -DE/DX = 0.0 !
! D4 D(3,1,16,5) 59.2306 -DE/DX = 0.0 !
! D5 D(3,1,16,9) -59.3255 -DE/DX = 0.0 !
! D6 D(3,1,16,12) 179.3912 -DE/DX = 0.0 !
! D7 D(4,1,16,5) 179.5783 -DE/DX = 0.0 !
! D8 D(4,1,16,9) 61.0223 -DE/DX = 0.0 !
! D9 D(4,1,16,12) -60.2611 -DE/DX = 0.0 !
! D10 D(6,5,16,1) 60.1308 -DE/DX = 0.0 !
! D11 D(6,5,16,9) -179.9966 -DE/DX = 0.0 !
! D12 D(6,5,16,12) -60.1212 -DE/DX = 0.0 !
! D13 D(7,5,16,1) 179.8544 -DE/DX = 0.0 !
! D14 D(7,5,16,9) -60.2729 -DE/DX = 0.0 !
! D15 D(7,5,16,12) 59.6025 -DE/DX = 0.0 !
! D16 D(8,5,16,1) -59.5944 -DE/DX = 0.0 !
! D17 D(8,5,16,9) 60.2783 -DE/DX = 0.0 !
! D18 D(8,5,16,12) -179.8463 -DE/DX = 0.0 !
! D19 D(10,9,16,1) 178.0724 -DE/DX = 0.0 !
! D20 D(10,9,16,5) 58.5807 -DE/DX = 0.0 !
! D21 D(10,9,16,12) -60.9147 -DE/DX = 0.0 !
! D22 D(11,9,16,1) 60.8869 -DE/DX = 0.0 !
! D23 D(11,9,16,5) -58.6048 -DE/DX = 0.0 !
! D24 D(11,9,16,12) -178.1002 -DE/DX = 0.0 !
! D25 D(17,9,16,1) -60.5202 -DE/DX = 0.0 !
! D26 D(17,9,16,5) 179.988 -DE/DX = 0.0 !
! D27 D(17,9,16,12) 60.4926 -DE/DX = 0.0 !
! D28 D(13,12,16,1) 60.2637 -DE/DX = 0.0 !
! D29 D(13,12,16,5) -179.5772 -DE/DX = 0.0 !
! D30 D(13,12,16,9) -61.0189 -DE/DX = 0.0 !
! D31 D(14,12,16,1) -179.3905 -DE/DX = 0.0 !
! D32 D(14,12,16,5) -59.2315 -DE/DX = 0.0 !
! D33 D(14,12,16,9) 59.3268 -DE/DX = 0.0 !
! D34 D(15,12,16,1) -59.7013 -DE/DX = 0.0 !
! D35 D(15,12,16,5) 60.4577 -DE/DX = 0.0 !
! D36 D(15,12,16,9) 179.016 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Everything is converged, therefore the optimisation has occurred correctly.
Frequency Analysis
Conducted an 'Optimisation and Frequency' analysis, the file can be found in the D-Space.
Summary Table:
| [N(CH3)3(CH2CN)]+ Optimisation and Frequency | Units | |
|---|---|---|
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| E(RB3LYP) | -306.3937605 | a.u. |
| RMS Gradient Norm | 0.00000046 | a.u. |
| Dipole Moment | 2.0625 | Debye |
| Point Group | C1 |
The calculations took 29 minutes, 15.1 seconds. The final total energy is -306.3937605, which is the same as the optimisation. This is correct since it means that the configuration did not change during the frequency analysis.
Item Table:
Item Value Threshold Converged?
Maximum Force 0.000002 0.000015 YES
RMS Force 0.000000 0.000010 YES
Maximum Displacement 0.000002 0.000060 YES
RMS Displacement 0.000001 0.000040 YES
Predicted change in Energy=-4.656264D-12
Optimization completed.
-- Stationary point found.
All the items are have been correctly converged.
Frequency Analysis:
From the .log file could find the 'low frequencies', which represent the minima of the cation.
Low frequencies --- -6.3125 -3.7644 -3.2824 -0.0014 -0.0011 -0.0009 Low frequencies --- 91.5807 153.8214 211.5421
The first line is in the accepted range for the frequencies, and the second line is not negative, therefore they fulfill the criteria.
IR Spectra:
Using the Gaussian Vibrational view, the IR spectra was calculated.
MO Analysis
Using the .fchk file from the optimisation, it was possible to perform a population analysis for the MO and NBO calculations.
Using the .fchk file after the Energy calculation, the MOs could be analysed. The file can be found in the D-Space.
The HOMO and LUMO orbitals could be illustrated with the results:
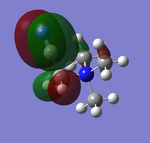

NBO Analysis
Using the .log file of the same calculations as the MO analysis, the charge dispersion and population analysis can be done.
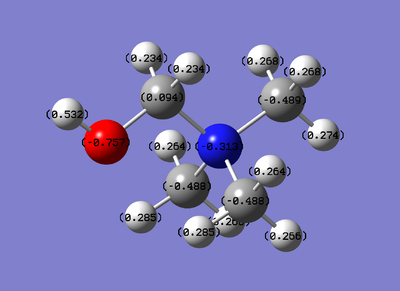
Using the same charge range as before (-1.125 to 1.125).
Qualitative view
Quantitative View
Observations:
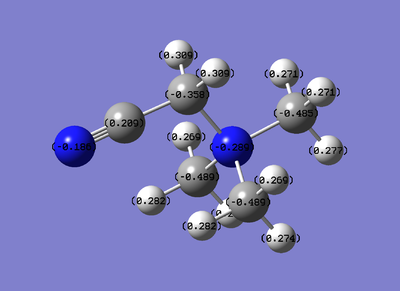
Viewing it in the quantitative view it is understandable why in the qualitative view none of the atoms have either a bright red or bright green colour as the range seems to be from -.489 to .309, therefore not to the extent of -1.125 or 1.125. It is worth noting that one of the three carbons that are not bonded to the -CN functional group has a different charge (-.358) compared to the other two (-.489). This explains why this interaction had a different bond angle compared to the other two carbons.
Comparisons of functional groups on charge distribution
Due to the strong negative charge on the oxygen, on can assume that the -OH functional group is electron donating to the central element (N). On the other hand, the -CN is just slightly electron withdrawing in this case (as the positive charge at the carbon is larger than the negative charge at the functional-group-nitrogen).
What the results show, is that when there is an electron donating functional group it increases the negative charge on the central element. On the opposite, electron withdrawing, makes it less-negative. The results also show that how much the central atom is changed is relative to how strongly electron donating/withdrawing the functional group is.
Comparisons of HOMO and LUMO
Can use the following table to observe the difference in between the HOMO and LUMOs as the functional group got changed. It would appear that the more electron withdrawing the functional group, the less of an energy gap it had.
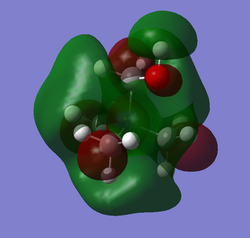
| HOMO | LUMO | LUMO energy | HOMO energy | Energy gap: | |
|---|---|---|---|---|---|
| [N(CH3)4]+ | 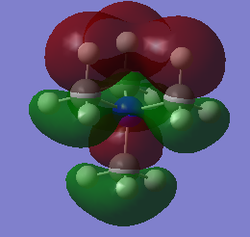
|
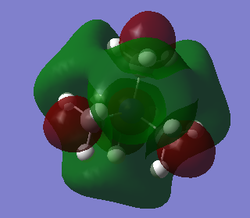
|
-0.13305 | -0.57929 | 0.44624 |
| [N(CH3)3(CH2OH)]+ | 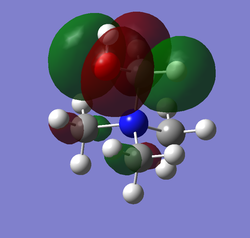
|
|
-0.11992 | -0.46628 | 0.34636 |
| [N(CH3)3(CH2CN)]+ | 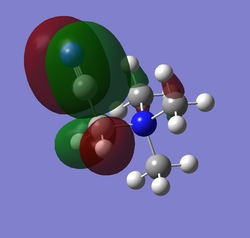
|
|
-0.18183 | -0.50048 | 0.31865 |