Rep:Mod:CSWmodule1 miniproject
Assigning Regioisomers in 'Click Chemistry'
Introduction
click chemistry is a chemical process that involves quickly and efficiently joining together small molecules. Click chemistry forms the basis for many reactions in nature and the term refers to a 'genre' of reactions rather than any specific reaction type. This mini project will consider a click chemistry reaction that can be catalysed by two different catalysts to give different regioisomers. Molecular modeling methods will be used to enable spectroscopic analysis of the isomers.
The 1,3-dipolar cycloaddition between an azide and an alkyne to give a 1,2,3-triazole
The reaction that will be studied in this project involves a 'click chemistry' reaction between a azide and an alkyne to give a 1,2,3-triazole.
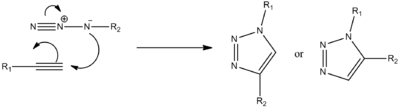
The reaction can be improved with use of a copper catalyst. (In 2002, two groups reported independently that the use of a Cu(I)-catalyst greatly speeds up the reaction.) Use of a Cu(I) catalyst however, leads to the predominant formation of isomer A. It has since been noted that use of a ruthenium (Rh(III)) catalyst leads to the predominant formation of isomer B. In this experiment the R groups will both be phenol groups. Reactions schemes for these reactions are shown below, in addition to 3D jmol representations of isomers A and B:
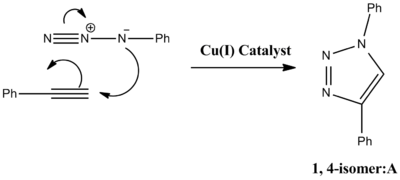
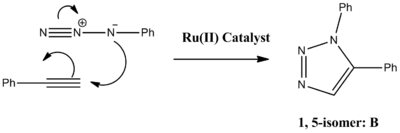
 |  |
The paper (J. Am. Chem. Soc. 2005, 127, 15998; DOI:10.1021/ja054114s ) discusses the 'Ruthenium-Catalyzed Clycloaddition of Alkynes and Organic Azides'. Using molecular modeling techniques to obtain spectroscopic data for isomers A and B, a comparison between the experimental data and the literature can be obtained.
This is done by creating a Gaussian Input File in 3DChemBio Ultra and minimising the energy using method DFT=mpw1pw91, then submitting the calculation to the shared SCAN server with instruction
"# mpw1pw91/6-31g(d,p) opt(maxcycle=25)."
A .gjf file containing the optimized geometry of the isomers can be obtained. Resubmission of the .gjf molecules (now with optimised geometry) under the instruction
"# mpw1pw91/6-31(d,p) NMR scrf(cpcm,solvent=chloroform)"
will create a .log file that can be opened in Gaussview5. In Gaussview 5, a NMR spectrum can then be viewed. The spectra are shown below:
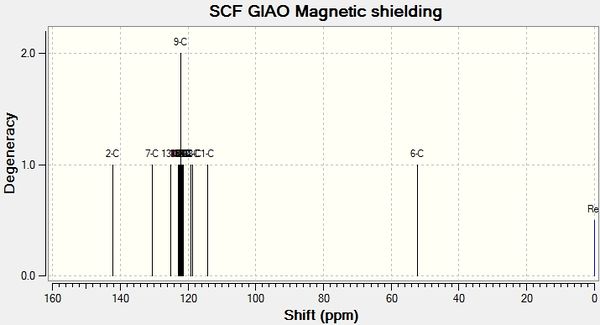
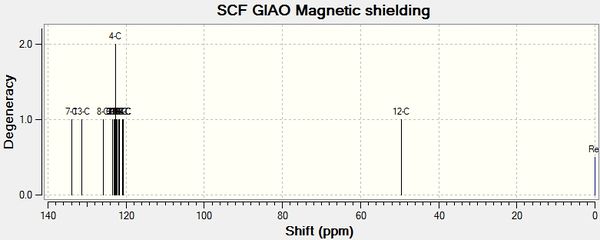
A comparison between the calculated NMR shift values and literature values can now be performed. The results have been tabulated below.
4.01| Isomer B Gaussian Shift ppm | Isomer B Literature Shift ppm [1] | Discrepancy |
| 49.51 | 51.85 | 2.34 |
| 121.95 | 126.93 | 4.98 |
| 122.34 | 127.22 | 4.88 |
| 122.46 | 128.22 | 5.76 |
| 122.75 | 128.92 | 6.17 |
| 122.89 | 129.08 | 6.19 |
| 123.14 | 129.64 | 6.5 |
| 123.48 | 133.26 | 9.78 |
| 125.86 | 133.34 | 7.48 |
| 131.65 | 135.66 | 4.01 |
| 133.94 | 138.26 | 4.32 |
| Isomer A Gaussian Shift ppm | Isomer A Literature Shift ppm | Discrepancy |
| 52.13 | 54.0 | 1.87 |
| 114.26 | 120.0 | 5.74 |
| 118.58 | 126.0 | 7.42 |
| 119.14 | 128.0 | 8.86 |
| 121.41 | 128.5 | 7.09 |
| 122.01 | 129.0 | 6.99 |
| 122.5 | 129.0 | 6.50 |
| 122.81 | 129.5 | 7.00 |
| 124.99 | 131.0 | 6.01 |
| 130.51 | 135.0 | 4.49 |
| 142.12 | 148.5 | 6.38 |
EXPLAINATION - ROOT MEAN SQUARE - GRAPH (BAR CHART) REFERENCES!!!
IR spectroscopy can also be helpful in analysing the two regioisomers A and B and reveal the physical differences between the two molecules.
This is done using a similar molecular modeling technique to the NMR analysis above. A Gaussian Input File in 3DChemBio Ultra and minimising the energy using method DFT=mpw1pw91, then submitting the calculation to the shared SCAN server with instruction
" # b3lyp/6-31G(d,p) opt freq"
This will create a .log file that can be opened in Gaussview5. In Gaussview 5, an IR spectrum can then be viewed. The spectra for isomers A and B are shown below:
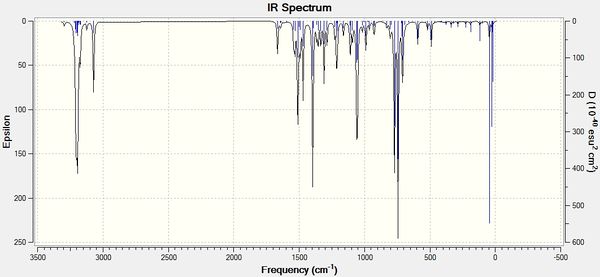
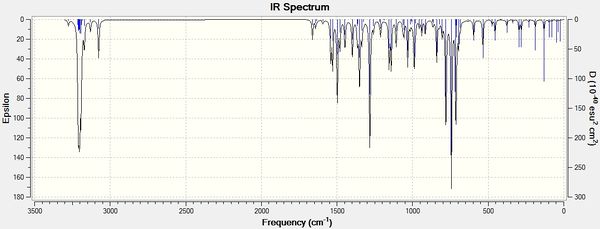
In the resulting output,
Sum of electronic and thermal Free Energies= -3170.440313
gives you in effect ΔG = ΔH - T.ΔS. You can use this term to compare the difference in free energies between two molecules, remembering that it is expressed in Hartrees; 1 Hartree = 627.5 kcal/mol.
[edit] Analyzing the Vibrational Spectrum
Download the.fchk file from the SCAN page, and by double-clicking, open it in Gaussview. From Results/Vibrations, select the Save normal modes from the Run FreqChk pop-up box and inspect the normal modes and their predicted intensities, using the animation feature to help describe them. Errors in the predicted wavenumbers are systematically too high for stretches (which means they can be corrected using empirical factors) by around 8%; bending and lower frequency modes are normally about right. Pay particular attention to the predicted intensities, which may help you to assign the vibrations. If you get any apparently negative modes, you will have in fact obtained a transition state (or higher order) stationary point.
- ↑ ↑ Supporting Info, J. Am. Chem. Soc. 2005, 127, 15998; DOI:10.1021/ja054114s
