Rep:Mod:CAI1234
NH3
Optimisation Data
An optimisation was run on an NH3 molecule, using the calculation method RB3LYP and basis set 6-31G(d,p). The final energy was found to be -56.55776873 a.u., the RMS gradient was 0.00000485 a.u. and the point group was C3V. The N-H bond distance was optimized at 1.01798 a.u. and the H-N-H bond angle was optimised at 105.741o. Literature values quote the bond length as 1.012A, and the bond angle as 106.7o[1]. There is a slight discrepancy between both the calculated N-H bond distance and H-N-H bond angle with the experimental literature values, suggesting the computational calculations are not ideal for this molecule. However, overall the error in both is less than 1%, which is a relatively successful prediction.
NH3 molecule |
The 'item' table from the results of the optimisation is presented below, showing the force and displacement have both been converged and the optimisation was completed correctly.
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986267D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The full log file is available below:
Media:CAI_NH3_OPTIMISE_text.LOG
Vibrational Modes
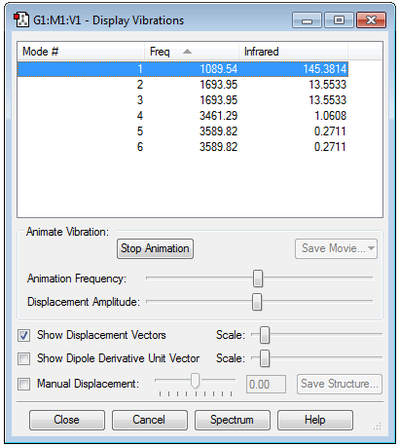
As ammonia contains four atoms and is a non-linear molecule, it is predicted that there will be 6 degrees of freedom: 3N-6=3(4)-6=6 degrees of freedom, and thus 6 vibrational modes are expected.
The generated vibrations show two sets of degenerate vibrational modes (i.e. of the same frequency) - modes 2 and 3, and modes 5 and 6.
Vibrational modes 1, 2 and 3 show N-H bond bending, and vibrational modes 4, 5 and 6 show N-H bond stretching. As expected, the stretching frequencies are higher than the bending frequencies.
Vibrational mode 4 is highly symmetric, and vibrational mode 1 is the umbrella mode.
4 bands are expected in the experimental spectrum.
Charge Distribution
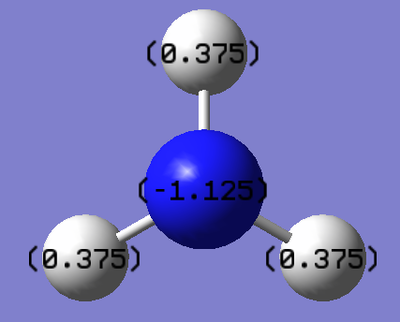
The charge on the nitrogen atom is found to be -1.125eV and the charge on the hydrogen atoms is found to be +0.375eV. A slightly polar covalent bond is expected in ammonia, due to the differing electronegativities of nitrogen and hydrogen. Nitrogen has an electronegativity of 3.04, whilst hydrogen has an electronegativity of 2.20[2] on the Pauling scale. Due to the greater electronegativity of nitrogen, this atom is expected to pull electron density towards itself, leading to a slight negative charge on the nitrogen atom, and an evenly split positive charge across hydrogen (as each hydrogen atom is in the same environment).
H2
Optimisation Data
An optimisation was run on an H2 molecule, using the calculation method RB3LYP and basis set 6-31G(d,p). The final energy was found to be -1.17853936 a.u., the RMS gradient was 0.00000017 a.u. and the point group was D*H. The H-H bond distance was optimized at 0.74279 a.u. and the H-H bond angle is 180o, as expected for a homonuclear diatomic molecule. Literature values quote the bond length as 0.74A[3], and thus the computed value is coherent with the literature value.
H2 molecule |
The 'item' table from the results of the optimisation is presented below, showing the force and displacement have both been converged and the optimisation was completed correctly.
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The full log file is available below:
Vibrational Modes

As H2 is a homonuclear diatomic molecule and thus is linear, it is predicted that there will be 1 degree of freedom: 3N-5=3(2)-5=1 vibrational mode, which is the result using GaussView. This vibrational mode is due to the H-H bond stretch.
N2
Optimisation Data
An optimisation was run on an N2 molecule, using the calculation method RB3LYP and basis set 6-31G(d,p). The final energy was found to be -109.52412868 a.u., the RMS gradient was 0.00000060 a.u. and the point group was D*H. The H-H bond distance was optimized at 1.10550 a.u. and the H-H bond angle is 180o, as expected for a homonuclear diatomic molecule. Literature values quote the bond length as 1.10A[3], and thus the computed value is coherent with the literature value.
N2 molecule |
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401052D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The full log file is available below:
Vibrational Modes
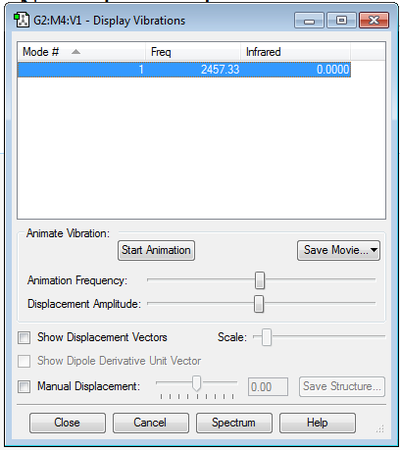
As with H2, N2 is a homonuclear diatomic molecule and thus is linear, it is predicted that there will be 1 degree of freedom: 3N-5=3(2)-5=1 vibrational mode, which is the result using GaussView. This vibrational mode is due to the H-H bond stretch.
Haber-Bosch Energy Calculation
The energies that have been calculated for H2, N2 and NH3 can be used to calculate a value for the energy change of the Haber-Bosch process.
3*H2 + N2 --> 2*NH3
E(NH3)=-56.55776873 a.u.
2*E(NH3)=-113.11553746 a.u.
E(N2)=-109.52412868 a.u.
E(H2)=-1.17853936 a.u.
3*E(H2)=-3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.0557907 a.u.
ΔE=2625.5*(-0.0557907)=-146.4784829 kJ.mol-1
The process is exothermic, and thus the products are more stable (deeper in energy) than the reactants.
The literature value for the enthalpy change of the Haber process is quoted as -91.8 kJ.mol-1[4], which is considerably lower than the computed value. The computational model has clearly failed here.
H2SiO
Optimisation Data
An optimisation was run on an H2SiO molecule, using the calculation method RB3LYP and basis set 6-31G(d,p). The final energy was found to be -365.90001403 a.u., the RMS gradient was 0.00000941 a.u. and the point group was CS. The Si=O bond distance was optimized at 1.53172 a.u. and the Si-H bond distance was optimized at 1.48652 a.u.. The O-Si-H bond angle was optimised at 124.156o, and the H-Si-H bond angle was optimised at 111.686o.
HSiO molecule |
Item Value Threshold Converged?
Maximum Force 0.000023 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000023 0.001800 YES
RMS Displacement 0.000017 0.001200 YES
Predicted change in Energy=-5.109774D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.4865 -DE/DX = 0.0 !
! R2 R(1,3) 1.4865 -DE/DX = 0.0 !
! R3 R(1,4) 1.5317 -DE/DX = 0.0 !
! A1 A(2,1,3) 111.686 -DE/DX = 0.0 !
! A2 A(2,1,4) 124.1565 -DE/DX = 0.0 !
! A3 A(3,1,4) 124.1576 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The full log file is available below:
Vibrational Modes
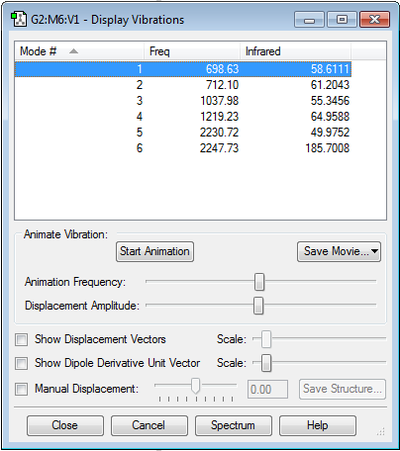
As H2SiO contains four atoms and is a non-linear molecule, it is predicted that there will be 6 degrees of freedom: 3N-6=3(4)-6=6 degrees of freedom, and thus 6 vibrational modes are expected.
The generated vibrations show no degenerate vibrational modes (i.e. of the same frequency).
Vibrational modes 1 and 2 show Si-H bond bending.
Vibrational modes 3 and 4 show bending of the Si-H bonds and stretching on the Si=O bond.
Vibrational modes 5 and 6 show Si-H bond stretching - with vibrational mode 5 showing a symmetric stretch and vibrational mode 6 showing an asymmetric stretch.
6 bands are expected in the experimental spectrum.
Charge Distributions
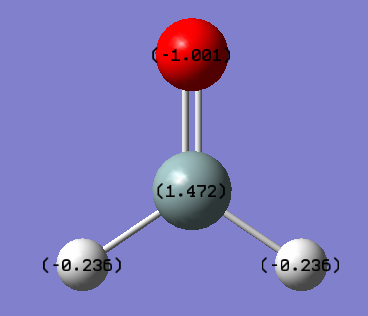
The charge on silicon was found to be +1.472 eV, the charge on oxygen was found to be -1.001 eV and the charge on hydrogen was found to be -0.236 eV. This follows the electronegativity difference between the elements. Silicon has the lowest electronegativity of 1.90[2] and thus attracts the electron density the least, leading to a positive charge on the silicon atom. Oxygen has the highest electronegativity of 3.44[2], and thus has the greatest negative charge as this atom attracts the electron density the most. Hydrogen has a slightly greater electronegativity than silicon at 2.20[2], and thus is slightly negatively charged with respect to the silicon.
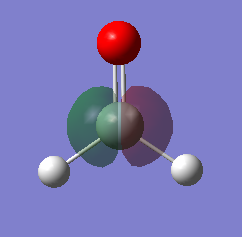
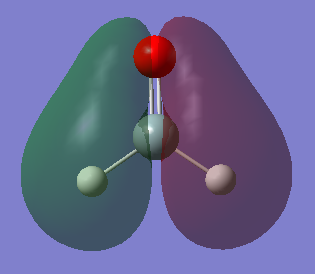
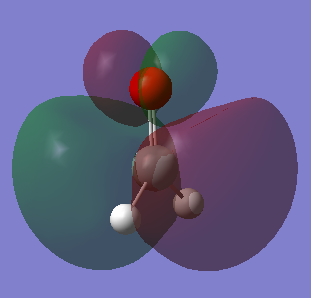
Molecular Orbitals
Five of the H2SiO molecular orbitals are analyzed below:


References
1: Ammonia Data Page, Wikipedia, retrieved 10/03/17, https://en.wikipedia.org/wiki/Ammonia_(data_page)
2: List of Electronegativity Values of the Elements, Science Notes, retrieved 10/03/17, http://sciencenotes.org/list-of-electronegativity-values-of-the-elements/
3: Common Bond Energies and Bond Lengths, Wired Chemist (Yoder, 2017), retrieved 10/03/17, http://www.wiredchemist.com/chemistry/data/bond_energies_lengths.html
4: Haber Process, Wikipedia, retrieved 10/03/17, https://en.wikipedia.org/wiki/Haber_process
