Rep:Mod:BC10151
NH3 Molecule
Optimisation
Optimised N-H bond distance: 1.01798 Å
Optimised H-N-H bond angle: 105.741
Calculation Method: B3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP) in atomic units (au):-56.55776873
RMS gradient:0.00000485
Point Group: C3v
NH3 |
The optimisation file is linked to here
Frequency Analysis
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES

How many modes do you expect from the 3N-6 rule? 3x4-6 = 6 modes
Which modes are degenerate (ie have the same energy)?
Modes 2 and 3 and modes 5 and 6 are degenerate.
Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending vibrations have a lower frequency than bending vibrations, which means that modes 1 to 3 are bending vibrations whereas 4 to 6 are stretching vibrations.
Which mode is highly symmetric?
Mode 4
One mode is known as the "umbrella" mode, which one is this?
Mode 1
How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
Degenerate modes would result in a single band thus there would only be 4 bands in the spectrum.
Population Analysis

Charge on the N-atom:-1.125
Charge on the H-atoms:0.375
The results fit with our predictions as we would expect N to have a negative value and H to have a positive value due to the fact that N is more electronegative than H and thus would pull the electron density towards itself, resulting in a more negative charge.
H2 Molecule
Optimisation
Optimised H-H bond distance: 0.60000 Å
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP) in atomic units (au): -1.17853936
RMS gradient:0.00000017 a.u.
Point Group:D∞h
H2 |
The optimisation file is linked to here
Frequency Analysis
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES

N2 Molecule
Optimisation
Optimised N-N bond distance:1.09200 Å
Calculation Method: RB3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP) in atomic units (au): -109.52412868
RMS gradient: 0.00000060 a.u.
Point Group:D∞h
N2 |
The optimisation file is linked to here
Frequency Analysis
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
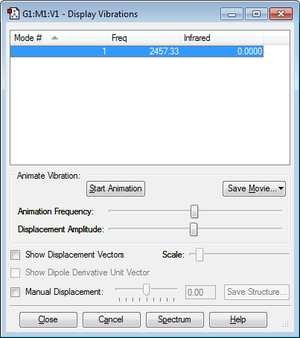
Determining the energy of the reaction of N2 + 3H2 -> 2NH3
E(NH3)= -56.55776873 au
2*E(NH3)= -113.1155375 au
E(N2)= -109.52412868 au
E(H2)= -1.17853936 au
3*E(H2)= -3.53561808 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579074 au
= -146.478599028 kJ/mol
We can therefore say that the ammonia product is more thermodynamically stable than the gaseous reactants as the reaction is exothermic so the product would be lower in energy than the reactants.
Project Molecule: HCl
Optimisation
Optimised H-Cl bond distance: 1.29000 Å
Calculation Method: B3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP) in atomic units (au): -460.80077875 a.u.
RMS gradient: 0.00005211 a.u.
Point Group:C∞v
HCl |
The optimisation file is linked to here
Frequency Analysis
Item Value Threshold Converged? Maximum Force 0.000090 0.000450 YES RMS Force 0.000090 0.000300 YES Maximum Displacement 0.000139 0.001800 YES RMS Displacement 0.000197 0.001200 YES
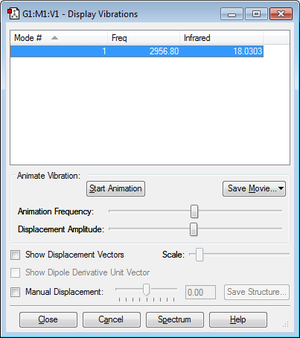
HCl has a single mode of vibration. This could be predicted using the 3N-5 rule, which we would use instead of the 3N-6 we used previously for NH3 because, as HCl is a linear molecule, we lose one axis of vibration. As we have 2 atoms N=2 therefore 3x2-5=1 mode. This mode corresponds to a symmetric stretching of the H-Cl bond, which would give a single band in the experimental spectrum of HCl.
The frequency of vibration of this bond could be precalculated using the following equation:
ν ˜ = 1/(2πc) √(k / μ) [1]
where ν ˜ is the wavenumber (which is the inverse of the frequency) k is the force constant (which relates to the bond strength), c is the speed of light and μ is the reduced mass, which can be calculated using the formula:
μ = m1m2/(m1+m2).
We can therefore determine that μ for HCl is 35/36, and taking k to be 481 N/m [2] , the following calculation can be carried out:
ν ˜ = 1/(2π*2.991x10^10) √[481 / (35/36)*1.66x10^-27] = 2953cm^-1, which is very similar to the value obtained from the frequency analysis.
Population Analysis

Charge on the Cl-atom:-0.284
Charge on the H-atom:0.284
As expected, Cl has a negative value due to the fact that it is more electronegative than H, pulling the electron density towards itself. The overall sum of charges is 0 as the molecule is neutral as a whole.
Studying the Molecular Orbitals of HCl





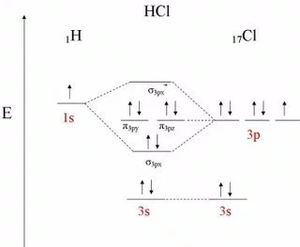
Cl2
Optimisation
Optimised Cl-Cl bond distance: 2.04174 Å
Calculation Method: B3LYP
Basis Set: 6-31G(d,p)
Final Energy E(RB3LYP) in atomic units (au): -920.34987886 a.u.
RMS gradient: 0.00002511 a.u.
Point Group:D∞h
Cl2 |
The optimisation file is linked to here
Frequency Analysis
Item Value Threshold Converged?
Maximum Force 0.000043 0.000450 YES
RMS Force 0.000043 0.000300 YES
Maximum Displacement 0.000121 0.001800 YES
RMS Displacement 0.000172 0.001200 YES
References
- ↑ This formula was taken from the Spectroscopy and Characterisation Lecture Notes on Infra-red Spectroscopy by Charlotte Williams.
- ↑ This is the reference for the bond constant of the H-Cl bond.
- ↑ This is the reference for the Molecular Diagram of HCl.
