Rep:Mod:Asaddat0987654321
Computational Chemistry Training
| BH3 3-21G | BH3 6-31G | GaBr3 | BBr3 | NH3 | |
|---|---|---|---|---|---|
| r(1-2) | 1.19467 | 1.19227 | 2.35018 | 1.93395 | 1.01798 |
| r(1-3) | 1.19445 | 1.19227 | 2.35018 | 1.93397 | 1.01798 |
| r(1-4) | 1.19480 | 1.19234 | 2.35018 | 1.93396 | 1.01798 |
| r(2-1-3) | 120.16 | 120.003 | 120.000 | 119.999 | 105.745 |
| r(2-1-4) | 119.986 | 119.994 | 120.000 | 120.000 | 105.745 |
| r(3-1-4) | 119.998 | 120.003 | 120.000 | 120.001 | 105.745 |
Week 1 Practice
Day 1 Calculations
09/11/2015
BH3 3-21G Optimization
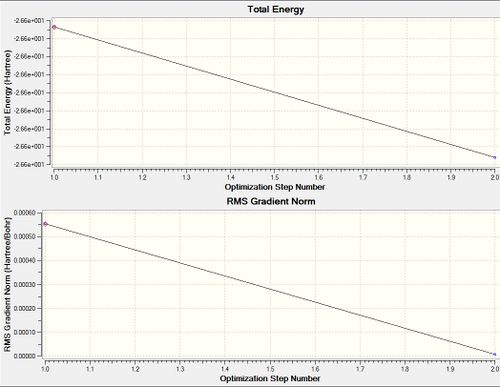
09112015 BH3 OPT File Name = 09112105_AS_BH3_321G_OPT File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 3-21G Charge = 0 Spin = Singlet E(RB3LYP) = -26.46226429 a.u. RMS Gradient Norm = 0.00008851 a.u. Imaginary Freq = Dipole Moment = 0.0003 Debye Point Group = CS Job cpu time: 0 days 0 hours 0 minutes 24.0 seconds.
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:09112105 AS BH3 321G OPT.LOG |
Item Value Threshold Converged? Maximum Force 0.000220 0.000450 YES RMS Force 0.000106 0.000300 YES Maximum Displacement 0.000940 0.001800 YES RMS Displacement 0.000447 0.001200 YES Predicted change in Energy=-1.672478D-07 Optimization completed. -- Stationary point found. |
|
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1948 -DE/DX = -0.0002 !
! R2 R(1,3) 1.1947 -DE/DX = -0.0002 !
! R3 R(1,4) 1.1944 -DE/DX = -0.0001 !
! A1 A(2,1,3) 120.0157 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.986 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9983 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Day 2 Calculations
09/11/2015
BH3 6-31G Optimization
09112015 BH3 OPT 631G File Name = 09112105_AS_BH3_631G_OPT File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -26.61532361 a.u. RMS Gradient Norm = 0.00000713 a.u. Imaginary Freq = Dipole Moment = 0.0001 Debye Point Group = CS Job cpu time: 0 days 0 hours 0 minutes 10.0 seconds.
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:09112105 AS BH3 631G OPT.LOG |
Item Value Threshold Converged?
Maximum Force 0.000012 0.000450 YES
RMS Force 0.000008 0.000300 YES
Maximum Displacement 0.000063 0.001800 YES
RMS Displacement 0.000039 0.001200 YES
Predicted change in Energy=-1.106101D-09
Optimization completed.
-- Stationary point found.
|
|
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0058 -DE/DX = 0.0 !
! A2 A(2,1,4) 119.9937 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0005 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
GaBr3 LAN2DZ Optimization
09112015 GABR3 OPT HPC FIRST TRIAL File Name = HPC_DLOAD_GABR3_09112015 File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = LANL2DZ Charge = 0 Spin = Singlet E(RB3LYP) = -41.70082770 a.u. RMS Gradient Norm = 0.00000016 a.u. Imaginary Freq = Dipole Moment = 0.0000 Debye Point Group = D3H Job cpu time: 0 days 0 hours 0 minutes 13.8 seconds.
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:HPC DLOAD GABR3 09112015.log |
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000003 0.001800 YES
RMS Displacement 0.000002 0.001200 YES
Predicted change in Energy=-1.307738D-12
Optimization completed.
-- Stationary point found.
|
|
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.3502 -DE/DX = 0.0 !
! R2 R(1,3) 2.3502 -DE/DX = 0.0 !
! R3 R(1,4) 2.3502 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
BBr3 6-31G Optimization
09112015 BBR3 OPT GEN File Name = 09112015_BBR3_HPC_OPT File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = Gen Charge = 0 Spin = Singlet E(RB3LYP) = -64.43644997 a.u. RMS Gradient Norm = 0.00000392 a.u. Imaginary Freq = Dipole Moment = 0.0001 Debye Point Group = CS Job cpu time: 0 days 0 hours 0 minutes 23.7 seconds.
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:09112015 BBR3 HPC OPT.log |
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000035 0.001800 YES
RMS Displacement 0.000024 0.001200 YES
Predicted change in Energy=-4.123635D-10
Optimization completed.
-- Stationary point found.
|
|
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0009 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0001 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9991 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Provide DSPACE Link
BH3 6-31G Frequency analysis
09112015 BH3 OPT 631G Frequency File Name = 09112105_AS_BH3_631G_Frequency File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -26.61532364 a.u. RMS Gradient Norm = 0.00000530 a.u. Imaginary Freq = 0 Dipole Moment = 0.0000 Debye Point Group = D3H Job cpu time: 0 days 0 hours 0 minutes 7.0 seconds.
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:09112105 AS BH3 631G FREQUENCY.LOG |
Item Value Threshold Converged?
Maximum Force 0.000011 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000042 0.001800 YES
RMS Displacement 0.000021 0.001200 YES
Predicted change in Energy=-6.630030D-10
Optimization completed.
-- Stationary point found.
|
|
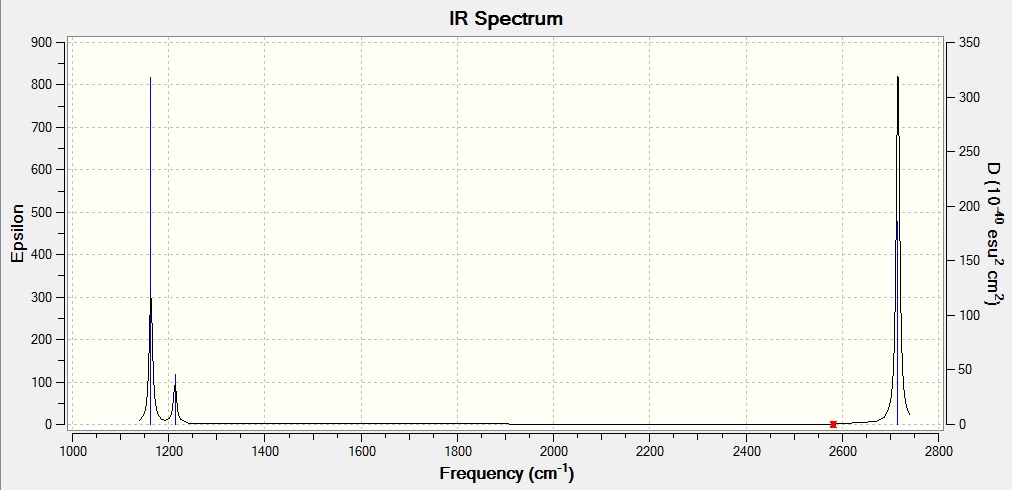
Low frequencies --- -14.5183 -14.5142 -10.8197 0.0003 0.0169 0.3454 Low frequencies --- 1162.9508 1213.1230 1213.1232
Wavenumber Intensity Infrared Type 1162.95 92.5706 Yes Bend 1213.12 14.0539 Yes Bend 1213.12 14.0533 No Bend 2582.66 0.0000 No Strech 2715.81 126.3291 No Strech 2715.81 126.3231 Yes Strech
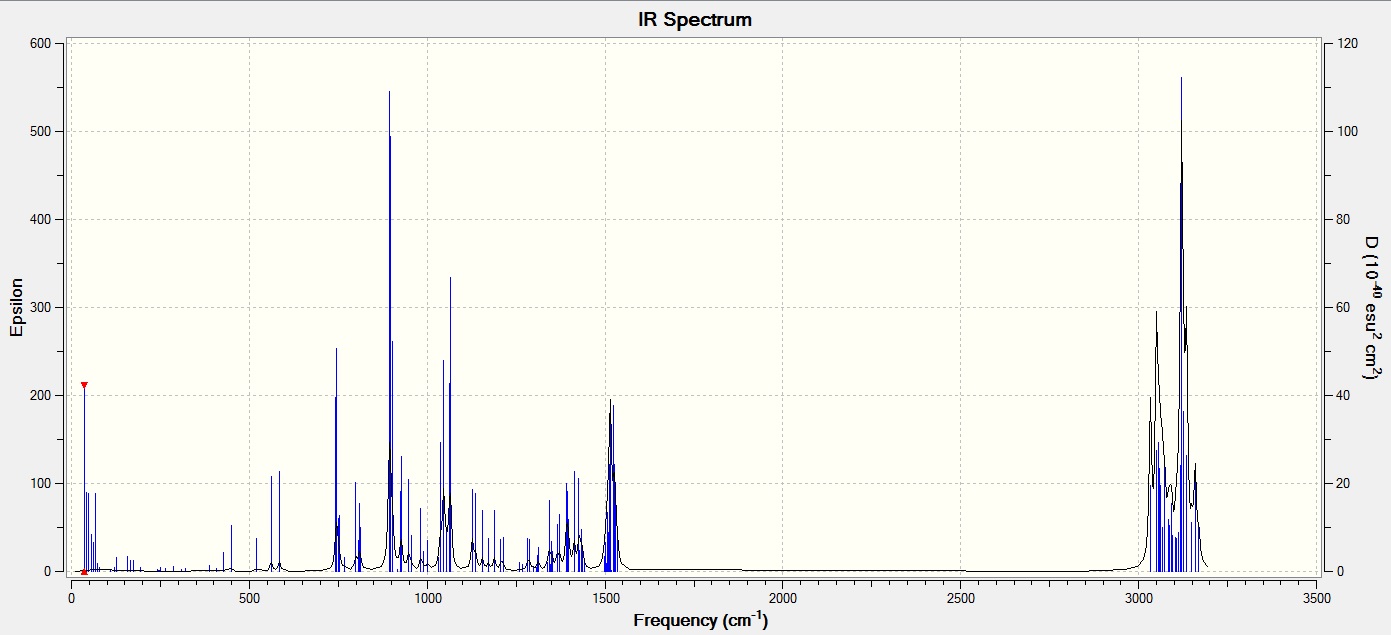
why are there 6 vibrations but only 3 peaks on the IR spectrum??
We know that for a given molecule the number of Vibration modes should be 3N - 6, where N is the number of atoms. In the case of BH3 since there are 4 atoms we get 6 vibration modes. As expected according to our Frequency analysis we find 6 vibrations modes which can be visualized. But, in the IR spectrum produced there are only 3 band peaks that we can observe. There are two reasons for this which have been detailed below.
1) According to the data there are two Vibrational modes at wavenumber 1213.12 cm-1 and 2715.81 cm-1. These have the same intensities and vibration frequency. Degenerate vibrations produce a single peak in the spectra because they have the same energy. Hence we see one peak for the two vibration frequency combined.
2) The intensity at the vibration frequency at 2582.66 cm-1 is a symmetrical stretch of BH3. And this frequency is inactive in the IR because this vibration produces no change in the dipole moment of the molecule. In order to be IR active, a vibration must cause a change in the dipole moment of the molecule.
GaBr3 LANL2DZ Frequency analysis
09112015 GABR3 Frequency analysis File Name = HPC_DLOAD_FAGABR3 File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = LANL2DZ Charge = 0 Spin = Singlet E(RB3LYP) = -41.70082770 a.u. RMS Gradient Norm = 0.00000025 a.u. Imaginary Freq = 0 Dipole Moment = 0.0000 Debye Point Group = C3H Job cpu time: 0 days 0 hours 0 minutes 12.4 seconds.
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:HPC DLOAD FAGABR3.log |
Item Value Threshold Converged?
Maximum Force 0.000011 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000042 0.001800 YES
RMS Displacement 0.000021 0.001200 YES
Predicted change in Energy=-6.630030D-10
Optimization completed.
-- Stationary point found.
|
|
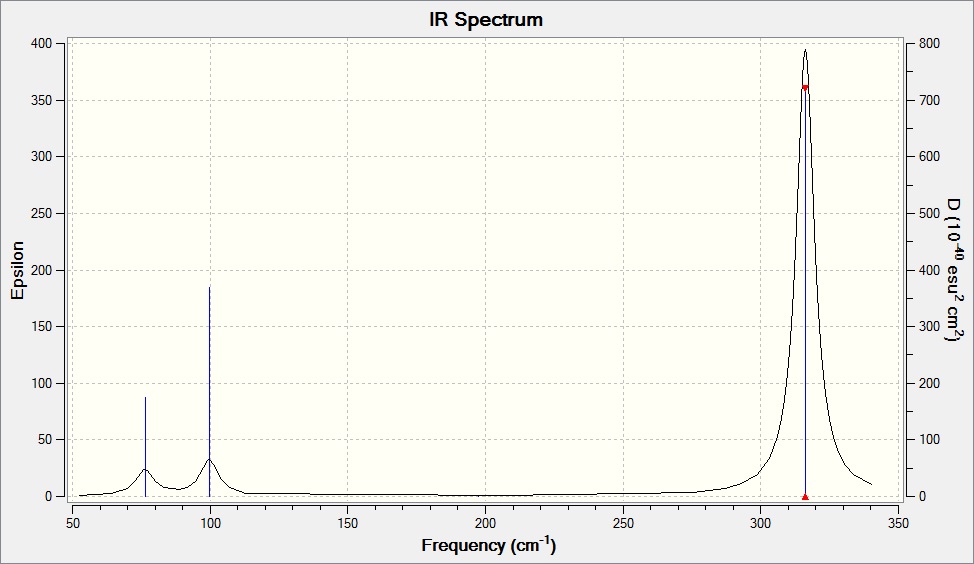
Low frequencies --- -1.4878 -0.0015 -0.0002 0.0096 0.6540 0.6540 Low frequencies --- 76.3920 76.3924 99.6767
Wavenumber Intensity Infrared Type 76.39 3.3451 Yes Bend 76.39 3.3450 no Bend 99.68 9.2166 Yes Bend 197.33 0.0000 no Stretch 316.18 57.0655 no Stretch 316.18 57.0669 Yes Stretch
What does the large difference in the value of the frequencies for BH3 compared to GaBr3 indicate?
The large difference in the frequencies indicate the difference in energy required to vibrate the BH3 molecule.
There been a reordering of modes! This can be seen particularly in relation to the A2" umbrella motion. Compare the relative frequency and intensity of the umbrella motion for the two molecules. Looking at the displacement vectors how has the nature of the vibration changed?
The GaBr3 molecule vibrates more along its displacement axis in the A2" symmetry compared to the GaBr3 molecule
Why?
Longer bond length.
Why must you use the same method and basis set for both the optimisation and frequency analysis calculations?
Basis sets determines the number of functions required to approximate the electronic structure of a certain molecule. An approximation made for a specific molecule needs to be compared to another molecule approximated using the exactly same method/basis set to keep the integrity of the values obtained. More importantly, when determining values like disassociation energy which needs to be calculated from values obtained from two different molecules but also the combined molecules, using different basis sets would lead to wrong values due to comparison of vastly different approximations.
What is the purpose of carrying out a frequency analysis?
The purpose of carrying out a frequency analysis is to figure out the minimum of our molecules potential energy surface. It helps figure out the vibration frequency which in turn helps predict Infrared spectroscopy peaks we might expect to see if the experiment was physically carried out.
What do the "Low frequencies" represent?
The low frequencies represent the motion of the center of mass of the molecule.
Molecular Orbitals of BH3
1st molecular orbital
2nd molecular orbital
3rd molecular orbital
Highest occupied molecular orbital
Lowest unoccupied molecular orbital
NH3 6-31G analysis
Summary :
NH3 molecule optimization File Name = 10112015_NH3_631G_OPT File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -56.55776873 a.u. RMS Gradient Norm = 0.00000323 a.u. Imaginary Freq = Dipole Moment = 1.8465 Debye Point Group = C3V Job cpu time: 0 days 0 hours 0 minutes 11.0 seconds.
Frequency Analysis Summary:
NH3 molecule frequency File Name = 10112015_NH3_631G_FREQ File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -56.55776872 a.u. RMS Gradient Norm = 0.00000322 a.u. Imaginary Freq = 0 Dipole Moment = 1.8465 Debye Point Group = C3 Job cpu time: 0 days 0 hours 0 minutes 7.0 seconds.
Population Analysis Summary:
NH3 molecule Population File Name = 10112015_NH3_631G_OPT File Type = .chk Calculation Type = SP Calculation Method = RB3LYP Basis Set = 6-31G(D,P) Charge = 0 Spin = Singlet Total Energy = -56.55776873 a.u. RMS Gradient Norm = 0.00000000 a.u. Imaginary Freq = Dipole Moment = 1.8465 Debye Point Group =
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:10112015 NH3 631G OPT.LOG |
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000012 0.001800 YES
RMS Displacement 0.000008 0.001200 YES
Predicted change in Energy=-9.844602D-11
Optimization completed.
-- Stationary point found.
|
|
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:10112015 NH3 631G FREQ Log.txt |
Item Value Threshold Converged?
Maximum Force 0.000006 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000013 0.001800 YES
RMS Displacement 0.000007 0.001200 YES
Predicted change in Energy=-1.131567D-10
Optimization completed.
-- Stationary point found.
|
|
Low frequencies --- -0.0138 -0.0026 -0.0009 7.0783 8.0932 8.0937 Low frequencies --- 1089.3840 1693.9368 1693.9368
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:10112015 NH3 631G POP LOG.txt |
N/A |
|
What are the specific NBO charges for the nitrogen and hydrogen atoms?
Ammonia-Borane Analysis
Optimization Summary
NH3BH3 Optimization File Name = 10112015 NH3BH3 631G OPT File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -83.22468893 a.u. RMS Gradient Norm = 0.00005974 a.u. Imaginary Freq = Dipole Moment = 5.5651 Debye Point Group = C1 Job cpu time: 0 days 0 hours 0 minutes 36.0 seconds.
Frequency Analysis Summary
NH3BH3 Frequency File Name = 10112015 NH3BH3 631G FREQ File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 0 Spin = Singlet E(RB3LYP) = -83.22468883 a.u. RMS Gradient Norm = 0.00005974 a.u. Imaginary Freq = 0 Dipole Moment = 5.5651 Debye Point Group = C1 Job cpu time: 0 days 0 hours 0 minutes 24.0 seconds.
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:10112015 NH3BH3 631G LOG.txt |
Item Value Threshold Converged?
Maximum Force 0.000123 0.000450 YES
RMS Force 0.000058 0.000300 YES
Maximum Displacement 0.000515 0.001800 YES
RMS Displacement 0.000296 0.001200 YES
Predicted change in Energy=-1.635696D-07
Optimization completed.
-- Stationary point found.
|
|
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:10112015 NH3BH3 631G FREQ LOG.txt |
Item Value Threshold Converged?
Maximum Force 0.000116 0.000450 YES
RMS Force 0.000060 0.000300 YES
Maximum Displacement 0.000581 0.001800 YES
RMS Displacement 0.000346 0.001200 YES
Predicted change in Energy=-1.740048D-07
Optimization completed.
-- Stationary point found.
|
|
E(NH3)= -56.55776873 au E(BH3)= -26.61532361 au E(NH3BH3)= -83.22468883 au
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)] ΔE = -83.22468883 –(-26.61532361)-( -56.55776873) ΔE = -0.05159649 au = 135.4666 Kj/mol
This is a relatively weak bond.
Tetra-Butyl Ammonium Ion Analysis
Optimization summary
N(C4H9)4+ Optimization File Name = QUATAMMONIUM_OPT File Type = .log Calculation Type = FOPT Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -685.98649942 a.u. RMS Gradient Norm = 0.00000128 a.u. Imaginary Freq = Dipole Moment = 1.2182 Debye Point Group = C1 Job cpu time: 0 days 3 hours 39 minutes 48.3 seconds.
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:QUATAMMONIUM OPT Log.txt |
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.001413 0.001800 YES
RMS Displacement 0.000197 0.001200 YES
Predicted change in Energy=-2.152879D-09
Optimization completed.
-- Stationary point found.
|
|
Frequency Analysis summary
N(C4H9)4+ Frequency File Name = QUATAMMONIUM_FREQ_LOG File Type = .log Calculation Type = FREQ Calculation Method = RB3LYP Basis Set = 6-31G(d,p) Charge = 1 Spin = Singlet E(RB3LYP) = -685.98649942 a.u. RMS Gradient Norm = 0.00000127 a.u. Imaginary Freq = 0 Dipole Moment = 1.2182 Debye Point Group = C1 Job cpu time: 0 days 4 hours 33 minutes 13.1 seconds.
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:QUATAMMONIUM FREQ LOG.log |
Item Value Threshold Converged? Maximum Force 0.000005 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.002534 0.001800 NO RMS Displacement 0.000482 0.001200 YES Predicted change in Energy=-3.711258D-09 |
|
Low frequencies --- -3.3879 -0.0010 -0.0007 0.0006 2.9807 4.1293 Low frequencies --- 35.3771 40.8237 46.9011
IR Spectrum Analysis
MO analysis
Optimisation log file here
| summary data | convergence | Jmol | |||
|---|---|---|---|---|---|
| File:QUATAMMONIUM MO LOG.log |
N/A |
|
HOMO
LUMO
Charge according to color
Charge numeric representation