Rep:Mod:AliceWickham
Bonding analyses using Ab initio and Density functional techniques
Optimisation of molecules
BH3
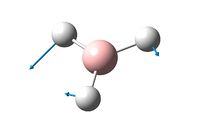
By assuming a given bond distance of 1.5A between the B and H nuclei, it was possible to solve the Schrodinger equation for the electrons finding the optimal bond lengths and bond angles for the BH3 molecule, using gaussview programming[1]. With results and constraints shown below.
From this programming it was shown that the optimal B-H bond length is 1.19A and that the optimal bond angle of the H-B-H bond is 120o. To ensure Gaussview had run to completion it was possible to check its file output and look at the forces and displacements, this information is shown below.
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004
! R2 R(1,3) 1.1935 -DE/DX = 0.0004
! R3 R(1,4) 1.1935 -DE/DX = 0.0004
! A1 A(2,1,3) 120.0 -DE/DX = 0.0
! A2 A(2,1,4) 120.0 -DE/DX = 0.0
! A3 A(3,1,4) 120.0 -DE/DX = 0.0
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Higher Basis Set
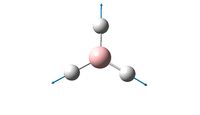
By using a higher basis set we can get a clearer and more accurate optimisaton of the BH3 molecule, this has been done below using a basis set of 6-31G which includes d and p orbitals.
Proof that it has run the optimisation to completion.
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
From this optimisation, it can be seen that the most stable B-H bond length is 1.19A and the optimal bond angle of H-B-H is still 120o.
TlBr3
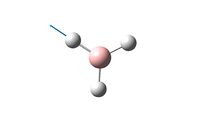
Having made the molecule of TlBr3 it was ran through the HPC scanner to find its optimal geometry.
The optimisation was run to completion as can be seen below:
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084037D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
From the optimisation it was shown that the most stable configuration has a Tl-Br bond length of 2.65A and a Br-Tl-Br bond angle of 120o. This bond length is similar to the value found in literature of 2.51A[2].
BBr3
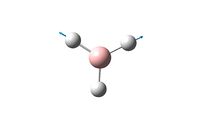
Otimisation of a BBr3 molecule was achieved on a HPC scanner starting from the optimised BH3 molecule prepared earlier.
It achieved completion as can be seen below;
Item Value Threshold Converged?
Maximum Force 0.000009 0.000450 YES
RMS Force 0.000006 0.000300 YES
Maximum Displacement 0.000047 0.001800 YES
RMS Displacement 0.000031 0.001200 YES
Predicted change in Energy=-6.331983D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.9511 -DE/DX = 0.0 !
! R2 R(1,3) 1.9511 -DE/DX = 0.0 !
! R3 R(1,4) 1.9511 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The optimised B-Br bond distance obtained from these calculations is 1.95A and the optimal Br-B-Br bond angle is 120o.
Comparison of the different molecules' bond lengths
| molecule | bond length |
|---|---|
| BH3 | 1.19A |
| TlBr3 | 2.65A |
| BBr3 | 1.95A |
As can be seen from the results the largest bond length is TlBr3 followed by BBr3 and BH3, with roughly equal differences between all 3. The differences in bond length can be attributed to two things; the nature of the ligand, and the nature of the central atom.
Changing the ligand from hydrogen to bromine appears to elongate the bond, this can be attributed to the different sizes of the ligands. As a smaller ligand hydrogen is able to get closer to the boron without feeling its repulsive forces and therefore its optimal structure is a tighter structure in total. The larger nature of the bromine means it has a greater number of electrons and therefore will have a higher repulsive force when at the H-B bond length. This requires the Br-B to be longer accommodating its larger size.
Looking at the difference between the two central atoms, (TlBr3 and BBr3) there is a larger bond length for the Thalium atom. Thalium and Boron are in the same group therefore have the same number of outer electrons and so would bond in the same way, the main difference between the two is the period in which they are in, Boron is in the 1st Thalium being in the 6th. This huge difference translates to the size of the central atom. A larger central atom has a larger electron density therefore requiring a larger area for the atom. This larger area means a ligand cannot get as close as it may be used to in smaller atoms, increasing the length of the bond.
A bond is a mutual attraction between atoms which allows formation of chemical substances mainly through the redistribution of electrons. This redistribution can occur in many ways leading to multiple types of bonds; covalent, ionic, metallic, to name but a few. As a result of these different redistributions bond will vary in strength according to the nature of their bonding, and bond lengths will also differ.
In Gaussview bonds do not always appear on the molecule in some images, even when the molecule is simple and known to have bonds. The lack of a bond shown on the image does not mean a bond is not there, more commonly the bond will not be shown in Gaussview until the atoms are at a distance where a bond is possible. If the atoms are too far away a bond will not show in Gaussview signifying there will be no bond in real life.
Frequency analysis
BH3
Taking the optimised BH3 with the higher basis set it was possible to analyze the frequencies of the molecule ensuring it is a minimum structure.
| Vibrational analysis of a BH3 molecule | |
|---|---|
| Programme constraints | |
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final Energy | -26.615au |
| Gradient | 0.000000237au |
| Dipole Moment | 0D |
| Point Group | D3H |
| Calculation Run Time | 10 seconds |
To check it completed;
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.323374D-10
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
frequencies found;
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824
Low frequencies --- 1163.0003 1213.1853 1213.1880
Animations can be looked at relating to the vibrational analysis of the molecule, these have been summarized below along with each vibrations frequency and its intensity.
The IR of the BH3 molecule can also be seen below, as noticed it does not have 6 peaks which could be expected due to the 6 different vibrational movements.
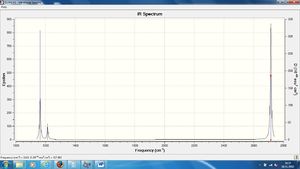
As can be seen there are only 3 peaks visible on the IR spectra, these are at frequencies of 1163, 1213, and 2715, (these values have been rounded up/down in the table) as would be expected. Although there are six vibrational motions, one of these has an intensity of 0 and therefore cannot be expected on the spectra as it will not be seen. Four of the motions can be split into pairs relating to their frequency (they have the same), this means a singular peak will be seen at this frequency, at an intensity which includes both motions. It can therefore be explained how 6 vibrational motions relates to 3 peaks on an IR spectra.
TlBr3
Having taken the optimised TlBr3 molecule made earlier it was possible to check it had optimised fully by looking at its vibrations, the main file used in this can be seen hereDOI:10042/21739 .
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | LAN2DZ |
| Energy | -91.218 a.u. |
| RMS Gradient Norm | 0.00000088 a.u. |
| Dipole Moment | 0D |
| Point Group | D3H |
| Run Time | 15.6 seconds |
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367
Low frequencies --- 46.4289 46.4292 52.1449
The lowest 'real' normal mode seen in these calculations is 46, as this is at least one order of magnitude larger than the highest '0' frequency (top line of the low frequencies) the optimisation has worked correctly. This is also backed up by the following,
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The IR spectrum obtained can be seen below, along with the frequencies and intensities.

| Frequency | Intensity |
|---|---|
| 50 | 4 |
| 50 | 4 |
| 50 | 6 |
| 170 | 0 |
| 210 | 25 |
| 210 | 25 |
Vibrational Analysis is carried out to ensure optimisation has completed and ensure that a minimum has been found, as the optimisation only tells if there is a stationary point. The low frequecies reperesent those that have been carried out at 0 wavelength. When doing optimisation adn frequency calculations the same basis set and method must be used otherwise this can affect structures and energies. Different basis sets give different energies.
Comparison of vibrational analysis
| BH3 | TlBr3 |
|---|---|
| 1160 | 46 |
| 1210 | 46 |
| 1210 | 52 |
| 2580 | 165 |
| 2720 | 211 |
| 2720 | 211 |
The most obvious difference between the two is the order of magnitude difference in the frequencies. This can be explained using Hooke's law, summarising the law, v (the frequency) is proportional to the root of k/μ. As k remains a constant the changeable factor in this is that of μ (the reduced mass of a molecule). As a result it can be seen that the mass of a molecule will invariably affect the frequency of its vibrations, in a negative way. A larger molecule will show smaller vibrational frequencies, due to the smaller energy required to vibrate the molecule. As well as the heavier nature of TlBr3, the longer bond it contains leads to a weaker bond which in turn leads to a lower vibrational frequency, due to the relationship between bond strength and vibration, show by the positive correlation between v and k, (classicly the mass, in this case the bond length). There has been a reordering of modes, with the A2" mode being higher in energy than the E' mode in TlBr3 however the same mode is lower in BBr3, however there are still the same number present. Both spectra show a singular peak at a higher frequency with two lower intensity peaks close together at a lower frequency, the TlBr3 spectra shows a broader two peaks at lower frequency than the sharpness in the BBr3 spectra. Other similarities are in both spectra there are two vibrational nodes close together and a further two close together higher in energy, this can be attributed to the way in which they vibrate, being either bending (lower) or stretching (higher).
Molecular Orbitals
BH3
MO analysis was carried out using the 6-31G(d,p) basis set, ensuring full NBOs was switched on and the full population analysis was carried out 'pop=full'. Using the optimised BH3 molecule we can look at the individual MO's which we can compare to our standard MO diagram. Link to the completed calculation can be found here DOI:10042/21666
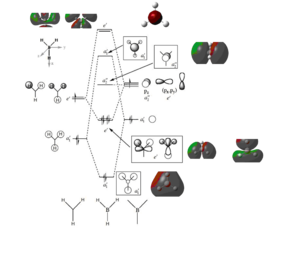
A molecular orbital diagram can be found on the right with the coresponding orbitals on it, those found via modelling in gaussview and those expected and drawn as common place LCAOs. Comparing the 'real' MOs which were generated by Gaussview and the LCAOs there does not seem to be a huge difference, apart from the number of electrons in the energy levels. The MOs contain 8 whereas the LCAOs only appear to contain 6, this would indicate the calculated MOs have an extra orbital energy level involved compared to the LCAOs seen. Both show nodes in the right places seperating the p orbitals into the specified charges. Due to the lack of dfference and the similarity shown in the LCAOs and the 'real' MOs, this is a positive thing for MO theory showing that it is not only accurate but can also be very useful in showing what to expect.
NBO Analysis of NH3
Before analysis can begin a molecule of NH3 was created, optimised checked by looking at the frequency and a population analysis was carried out.
Optimisation
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -56.557 a.u. |
| RMS Gradient Norm | 0.00000289 a.u. |
| Dipole Moment | 1.85D |
| Point Group | C3V |
| Run Time | 20 seconds |
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000010 0.001800 YES
RMS Displacement 0.000007 0.001200 YES
Predicted change in Energy=-7.826471D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7463 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7463 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7463 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 111.867 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
From the optimisation it is seen that the optimal N-H bond length is 1.02A, and the optimal H-N-H bond angle is 105.7o.
Frequency Analysis
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -56.557 a.u. |
| RMS Gradient Norm | 0.00000138 a.u. |
| Dipole Moment | 1.85D |
| Point Group | C3V |
| Run Time | 16 seconds |
Item Value Threshold Converged?
Maximum Force 0.000004 0.000015 YES
RMS Force 0.000001 0.000010 YES
Maximum Displacement 0.000007 0.000060 YES
RMS Displacement 0.000004 0.000040 YES
Predicted change in Energy=-1.900110D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7447 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7444 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7443 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8637 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies --- -9.3372 -8.1982 -6.2791 -0.0015 -0.0014 -0.0012
Low frequencies --- 1089.3347 1693.9209 1693.9246
1 2 3
A A A
Frequencies -- 1089.3347 1693.9209 1693.9246
As expected there are six '0' frequency values, corresponding to the 3N-6 rule, these frequencies are all at least one magnitude of 10 smaller than the actual frequencies observed. The three frequencies seen here are all positive further showing the calculation was successful.
MO analysis
Also included are the molecular orbitals found at the individual MO'S.
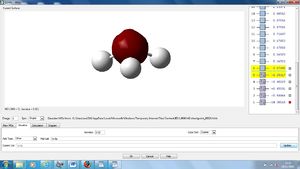
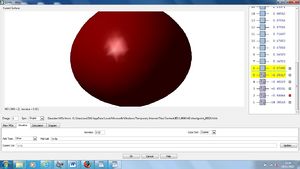
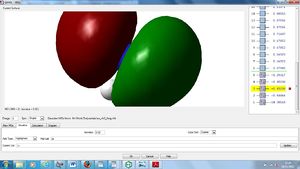
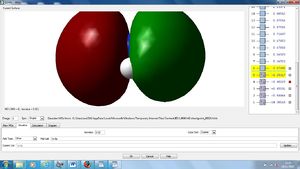
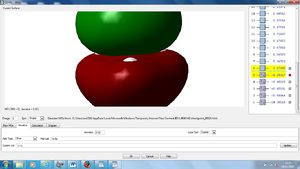
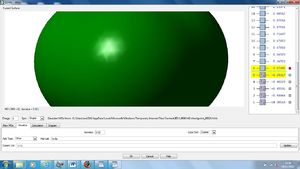
NBO Analysis
Using our optimised NH3 molecule a Natural Bond Orbital Analysis was carried out, looking at the charges placed on the molecules, by colour coordination and using actual numbers.
| Atom | Charge |
|---|---|
| Nitrogen | -1.13D |
| Hydrogen | 0.33D |
As expected this shows nitrogen has a high electron density, due to its high electronegativity and therefore ability to withdraw electrons from the hydrogen leaving them with positive charges.
Other data collected using NBO analysis is shown below,
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
-0.0001 -0.4986 -0.0059 0.0000 -0.2910
0.0052 0.8155 0.0277 0.0000 0.0000
0.0281 0.0000 0.0000 0.0032 0.0082
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0000 0.0072 -0.0289 0.0000
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4077 0.0138 0.7062 0.0240
0.0140 0.0243 0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 -0.0250
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2909
-0.0052 0.4077 0.0138 -0.7062 -0.0239
0.0140 -0.0243 -0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 0.0250
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 -0.8618
0.0505 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0269 0.0155
6. (0.00000) RY*( 1) N 1 s( 99.98%)p 0.00( 0.02%)d 0.00( 0.00%)
7. (0.00000) RY*( 2) N 1 s(100.00%)
8. (0.00000) RY*( 3) N 1 s( 0.03%)p99.99( 99.97%)d 0.01( 0.00%)
9. (0.00000) RY*( 4) N 1 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
10. (0.00000) RY*( 5) N 1 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%)
11. (0.00000) RY*( 6) N 1 s( 0.00%)p 1.00( 0.12%)d99.99( 99.88%)
12. (0.00000) RY*( 7) N 1 s( 0.00%)p 1.00( 0.12%)d99.99( 99.88%)
13. (0.00000) RY*( 8) N 1 s( 0.00%)p 1.00( 0.01%)d99.99( 99.99%)
14. (0.00000) RY*( 9) N 1 s( 0.01%)p 4.42( 0.06%)d99.99( 99.92%)
15. (0.00000) RY*(10) N 1 s( 0.00%)p 1.00( 0.03%)d99.99( 99.97%)
16. (0.00112) RY*( 1) H 2 s( 72.77%)p 0.37( 27.23%)
0.0038 0.8531 0.5218 -0.0017 -0.0001
17. (0.00045) RY*( 2) H 2 s( 26.59%)p 2.76( 73.41%)
-0.0017 0.5157 -0.8435 -0.1501 -0.0007
18. (0.00034) RY*( 3) H 2 s( 0.00%)p 1.00(100.00%)
0.0000 0.0004 -0.0005 -0.0001 1.0000
19. (0.00000) RY*( 4) H 2 s( 0.72%)p99.99( 99.28%)
20. (0.00112) RY*( 1) H 3 s( 72.77%)p 0.37( 27.23%)
0.0038 0.8531 0.5218 0.0009 0.0014
21. (0.00045) RY*( 2) H 3 s( 26.59%)p 2.76( 73.41%)
-0.0017 0.5157 -0.8435 0.0756 0.1297
22. (0.00034) RY*( 3) H 3 s( 0.00%)p 1.00(100.00%)
0.0000 0.0004 -0.0005 -0.8660 0.5000
23. (0.00000) RY*( 4) H 3 s( 0.72%)p99.99( 99.28%)
24. (0.00112) RY*( 1) H 4 s( 72.76%)p 0.37( 27.24%)
0.0038 0.8530 0.5219 0.0009 -0.0015
25. (0.00045) RY*( 2) H 4 s( 26.61%)p 2.76( 73.39%)
-0.0017 0.5158 -0.8435 0.0750 -0.1300
26. (0.00034) RY*( 3) H 4 s( 0.00%)p 1.00(100.00%)
0.0000 0.0000 0.0000 0.8660 0.5000
27. (0.00000) RY*( 4) H 4 s( 0.72%)p99.99( 99.28%)
28. (0.00000) BD*( 1) N 1 - H 2
( 31.17%) 0.5583* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
( 68.83%) -0.8297* H 2 s( 99.91%)p 0.00( 0.09%)
29. (0.00000) BD*( 1) N 1 - H 3
( 31.17%) 0.5583* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
( 68.83%) -0.8297* H 3 s( 99.91%)p 0.00( 0.09%)
30. (0.00000) BD*( 1) N 1 - H 4
( 31.17%) 0.5583* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
( 68.83%) -0.8297* H 4 s( 99.91%)p 0.00( 0.09%)
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12515 1.99982 6.11104 0.01429 8.12515
H 2 0.37505 0.00000 0.62250 0.00246 0.62495
H 3 0.37505 0.00000 0.62250 0.00246 0.62495
H 4 0.37505 0.00000 0.62249 0.00246 0.62495
=======================================================================
* Total * 0.00000 1.99982 7.97852 0.02166 10.00000
Summarissing the NBO analysis the Nitrogen has a charge of -1.125D and each individual Hydrogen has a charge of .375D. All calculations were carried out on HPC scanner and can be seen via the following link DOI:10042/21722
Energies
A molecule of NH3BH3 was created starting with a molecule of ethylene, and changing the carbon atoms for a nitrogen and a boron.
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | UB3LYP |
| Basis Set | 6-31G(d,p) |
| E(UB3LYP) | -82.57 a.u. |
| RMS Gradient Norm | 0.00010609 a.u. |
| Dipole Moment | 3.23D |
| Point Group | C1 |
| Job cpu time | 0 days 0 hours 2 minutes 25.9 seconds |
As it was run with a no symm constaint the summary shows the point group to be C1 whereas infact it is C3v.
Item Value Threshold Converged?
Maximum Force 0.000199 0.000450 YES
RMS Force 0.000071 0.000300 YES
Maximum Displacement 0.000887 0.001800 YES
RMS Displacement 0.000384 0.001200 YES
Predicted change in Energy=-3.298934D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,4) 1.2834 -DE/DX = -0.0002 !
! R2 R(2,4) 1.2097 -DE/DX = 0.0001 !
! R3 R(3,4) 1.2097 -DE/DX = 0.0001 !
! R4 R(4,5) 1.4677 -DE/DX = 0.0001 !
! R5 R(5,6) 1.0133 -DE/DX = -0.0001 !
! R6 R(5,7) 1.0133 -DE/DX = -0.0001 !
! A1 A(1,4,2) 101.6665 -DE/DX = 0.0 !
! A2 A(1,4,3) 101.6509 -DE/DX = 0.0 !
! A3 A(1,4,5) 103.8288 -DE/DX = 0.0 !
! A4 A(2,4,3) 120.8454 -DE/DX = 0.0001 !
! A5 A(2,4,5) 112.8925 -DE/DX = -0.0001 !
! A6 A(3,4,5) 112.8897 -DE/DX = -0.0001 !
! A7 A(4,5,6) 123.267 -DE/DX = 0.0 !
! A8 A(4,5,7) 123.266 -DE/DX = 0.0 !
! A9 A(6,5,7) 113.2122 -DE/DX = 0.0 !
! D1 D(1,4,5,6) -86.9218 -DE/DX = 0.0 !
! D2 D(1,4,5,7) 86.8833 -DE/DX = 0.0 !
! D3 D(2,4,5,6) 22.3468 -DE/DX = 0.0 !
! D4 D(2,4,5,7) -163.8481 -DE/DX = 0.0 !
! D5 D(3,4,5,6) 163.8288 -DE/DX = 0.0 !
! D6 D(3,4,5,7) -22.3661 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Vibrational Analysis was then carried out,
Item Value Threshold Converged?
Maximum Force 0.000315 0.000450 YES
RMS Force 0.000104 0.000300 YES
Maximum Displacement 0.001590 0.001800 YES
RMS Displacement 0.000561 0.001200 YES
Predicted change in Energy=-4.624683D-07
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies --- 0.0012 0.0013 0.0013 16.4356 16.9403 36.7832 Low frequencies --- 265.7790 632.2050 639.2800
1 2 3
A A A
Frequencies -- 265.7789 632.2050 639.2800
As can be seen the optimisation has run to completion, can be seen in both the item section of optimisation and in the vibrational part, as there are no negative frequencies it can also be seen that the calculation has run correctly.
The optimal bond length, B-H 1.02A, N-H 1.21A, N-B 1.67A Optimal bond angles, H-N-H 113.87°, H-B-H 107.87°
The association energies can now be read and calculated,
| E[NH3] | -56.55776856 |
| E[BH3] | -26.61532252 |
| E[NH3BH3] | -83.22468956 |
| ΔE | 0.05159848 |
This gives us the association energy of combining a molecule of NH3 and BH3 in au, in kJ/mol this would be, -135.51kJ/mol. This is the dissociation energy of the N-B bond, and is similar to literature values.
Investigating Aromaticity
Benzene is a widely known molecule having a typical conjugated system, its for this reason that benzene will be the reference molecule when looking at vibrations and MOs of 3 other molecules, boratabenzene, pyridinium and borazine.
Benzene
Looking at a molecule of benzene as a reference in looking at aromaticity. To start this a molecule of benzene was made and optimised on a HPC scanner with a 6-31G(d,p) basis set, results can be found here DOI:10042/21825
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -232.258 a.u. |
| RMS Gradient Norm | 0.00009549 a.u. |
| Dipole Moment | 0D |
| Point Group | D6h |
| Run Time | 1 minute 56.4 seconds |
Item Value Threshold Converged?
Maximum Force 0.000212 0.000450 YES
RMS Force 0.000085 0.000300 YES
Maximum Displacement 0.000991 0.001800 YES
RMS Displacement 0.000315 0.001200 YES
Predicted change in Energy=-5.157454D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0001 !
! R2 R(1,6) 1.3961 -DE/DX = 0.0002 !
! R3 R(1,7) 1.0861 -DE/DX = 0.0002 !
! R4 R(2,3) 1.3961 -DE/DX = 0.0002 !
! R5 R(2,8) 1.0861 -DE/DX = 0.0002 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0001 !
! R7 R(3,9) 1.086 -DE/DX = 0.0002 !
! R8 R(4,5) 1.3961 -DE/DX = 0.0002 !
! R9 R(4,10) 1.086 -DE/DX = 0.0002 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0861 -DE/DX = 0.0002 !
! R12 R(6,12) 1.0861 -DE/DX = 0.0002 !
! A1 A(2,1,6) 119.9972 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.9949 -DE/DX = 0.0 !
! A3 A(6,1,7) 120.0079 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0079 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.9881 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.004 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.9948 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0086 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9966 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.9972 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9934 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0094 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.0083 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0014 -DE/DX = 0.0 !
! A15 A(6,5,11) 119.9904 -DE/DX = 0.0 !
! A16 A(1,6,5) 119.9946 -DE/DX = 0.0 !
! A17 A(1,6,12) 120.0106 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9948 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.0059 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 180.0023 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -180.01 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0019 -DE/DX = 0.0 !
! D5 D(2,1,6,5) -0.0055 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.9972 -DE/DX = 0.0 !
! D7 D(7,1,6,5) -180.0013 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.007 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0117 -DE/DX = 0.0 !
! D10 D(1,2,3,9) -179.9914 -DE/DX = 0.0 !
! D11 D(8,2,3,4) 180.0036 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0005 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0062 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0059 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9969 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0028 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.0051 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 180.0058 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -180.0055 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.0054 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.011 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 180.0027 -DE/DX = 0.0 !
! D23 D(11,5,6,1) -179.9999 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0082 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
We also used frequency analysis to confirm the optimisation ran to completion, DOI:10042/21826
Item Value Threshold Converged?
Maximum Force 0.000190 0.000450 YES
RMS Force 0.000092 0.000300 YES
Maximum Displacement 0.000971 0.001800 YES
RMS Displacement 0.000372 0.001200 YES
Predicted change in Energy=-4.896874D-07
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies --- -15.5663 -14.5022 -13.9318 -0.0006 -0.0005 0.0006
Low frequencies --- 414.0684 414.1453 620.9624
As there are no negative frequencies present the calculation has worked and it can be accepted that the minimum energy state has been found.
1 2 3
A A A
Frequencies -- 414.0683 414.1453 620.9624
Shown are the frequencies of the peaks on the IR spectra (also shown) and their intensities.
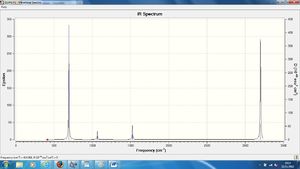
| No. | Frequency | Intensity | No. | Frequency | Intensity | No. | Frequency | Intensity |
|---|---|---|---|---|---|---|---|---|
| 1 | 414.07 | 0 | 11 | 1012.37 | 0 | 21 | 1524.35 | 6.62 |
| 2 | 414.15 | 0 | 12 | 1017.88 | 0 | 22 | 1524.43 | 6.61 |
| 3 | 620.96 | 0 | 13 | 1019.95 | 0 | 23 | 1653.03 | 0 |
| 4 | 620.97 | 0 | 14 | 1066.34 | 3.40 | 24 | 1653.15 | 0 |
| 5 | 693.05 | 74.25 | 15 | 1066.43 | 3.40 | 25 | 3174.34 | 0 |
| 6 | 718.31 | 0 | 16 | 1179.25 | 0 | 26 | 3183.81 | 0 |
| 7 | 864.10 | 0 | 17 | 1202.17 | 0 | 27 | 3183.99 | 0 |
| 8 | 864.17 | 0 | 18 | 1202.28 | 0 | 28 | 3199.48 | 46.58 |
| 9 | 973.64 | 0 | 19 | 1356.09 | 0 | 29 | 3199.65 | 46.52 |
| 30 | 3210.15 | 0 |
From all these calculations the optimal C-C bond length in benzene is 1.396A, and the optimal C-C-C bond angle is 120o. These bond lengths are as would be expected in benzene. They are somewhere between average C-C single and double bond lengths notably due to the conjugation of bonds in the molecule. The bond angle is also to be expected due to the sp2 hybridisation of the carbon atoms in the molecule
Having found the optimal geometry of benzene we used the software to look at the MO'S of benzene and use this to construct the LCAOs. The completed LCAOs in their relative energies can be seen below, with raw data found here DOI:10042/21845
We were then able to carry out NBO analysis.
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.23852 1.99910 4.22611 0.01331 6.23852
C 2 -0.23855 1.99910 4.22613 0.01331 6.23855
C 3 -0.23854 1.99910 4.22613 0.01331 6.23854
C 4 -0.23852 1.99910 4.22611 0.01331 6.23852
C 5 -0.23855 1.99910 4.22613 0.01331 6.23855
C 6 -0.23854 1.99910 4.22613 0.01331 6.23854
H 7 0.23854 0.00000 0.76003 0.00144 0.76146
H 8 0.23853 0.00000 0.76003 0.00144 0.76147
H 9 0.23854 0.00000 0.76002 0.00144 0.76146
H 10 0.23854 0.00000 0.76003 0.00144 0.76146
H 11 0.23853 0.00000 0.76003 0.00144 0.76147
H 12 0.23854 0.00000 0.76002 0.00144 0.76146
=======================================================================
* Total * 0.00000 11.99462 29.91690 0.08847 42.00000
Natural Population
--------------------------------------------------------
Core 11.99462 ( 99.9552% of 12)
Valence 29.91690 ( 99.7230% of 30)
Natural Minimal Basis 41.91153 ( 99.7893% of 42)
Natural Rydberg Basis 0.08847 ( 0.2107% of 42)
--------------------------------------------------------
Atom No Natural Electron Configuration
----------------------------------------------------------------------------
C 1 [core]2S( 0.96)2p( 3.26)3p( 0.01)
C 2 [core]2S( 0.96)2p( 3.26)3p( 0.01)
C 3 [core]2S( 0.96)2p( 3.26)3p( 0.01)
C 4 [core]2S( 0.96)2p( 3.26)3p( 0.01)
C 5 [core]2S( 0.96)2p( 3.26)3p( 0.01)
C 6 [core]2S( 0.96)2p( 3.26)3p( 0.01)
H 7 1S( 0.76)
H 8 1S( 0.76)
H 9 1S( 0.76)
H 10 1S( 0.76)
H 11 1S( 0.76)
H 12 1S( 0.76)
Interpreting the NBO data tells us that each Carbon contributes 50% to each C-C bond, and each carbon contributes 62.04% in a C-H bond with the hydrogen contributing the remaining 37.96%. This tells us that the C-C bond is a far more equal character bond than that of the C-H.NBO analysis can also show us the sp character of the carbon and hydrogen atoms in the benzene molecule. These have been tabulated below,
| Bond | Coefficient | Hybridisation |
|---|---|---|
| C1-C2 | 0.7071, 0.7071 | s(35.19%) p(64.77%) both |
| C1-C6 | 0.7071, 0.7071 | s(35.19%) p(64.77%) both |
| C1-C6 | 0.7071, 0.7071 | s(0%) p(99.96% both |
| C2-C3 | 0.7071, 0.7071 | s(35.21%) p(64.76%) both |
| C2-C3 | 0.7071, 0.7071 | s(0%) p(99.96%) both |
| C3-C4 | 0.7071, 0.7071 | s(35.19%) p(64.77%) both |
| C4-C5 | 0.7071, 0.7071 | s(35.20%) p(64.76%) both |
| C4-C5 | 0.7071, 0.7071 | s(0%) p(99.96%) both |
| C5-C6 | 0.7071, 0.7071 | s(35.20%) p(64.76%) both |
| C-H | 0.7876, 0.6161 | C:s(29.57%) p(70.39%), H:s(99.95%) p(0.05%) |
As can be seen the carbon atoms in a C-C bond show values corresponding to sp2 hybridization which would be expected of a carbon with 3 sigma bonds. The second bond seen between the 2 carbons can be seen to be pi as from the data they contain no s orbital character. All C-H bonds in benzene show the same orbital character with carbon showing sp2 hybridized properties with further emphasis on the p orbitals of the carbon atom. As expected the hydrogen atoms have s character only. NBO analysis can also be used to determine the charge distribution in a molecule, and this was carried out, showing the charges by colour diferences and also showing the actual dipole values recorded. These can be seen in the pictures below. As noted the red corresponds to a dipole of -0.239 and the green corresponds to +0.239.
As can be seen the charge distribution is as would be expected, due to the lack of electron withdrawing groups being present.
| Atom | Charge |
|---|---|
| Carbon | -0.239D |
| Hydrogen | +0.239D |
Aromaticity
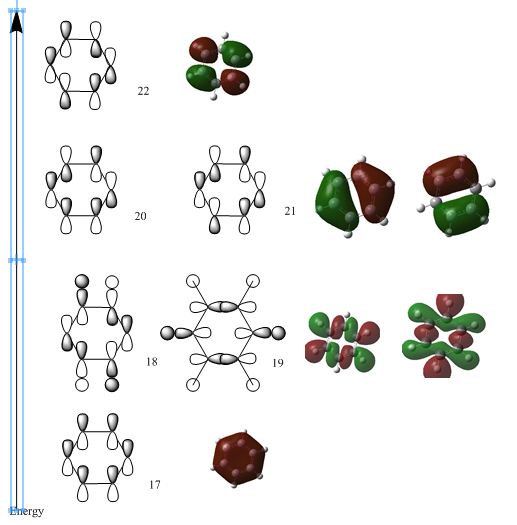
Aromaticity is a property which describes how a conjugated ring can exhibit stabilization properties stronger than those that would be expected by stailisation alone. The cyclic molecules to which this applies have a 4n+2 electron count, which lower the energy of the molecule. Benzene has this 4n+2 electron count, where n=1, the cyclic nature can be seen. Cyclic conjugation and therefore aromaticity, is only related to pi orbitals, helping understand why in MO diagrams of benzene the sigma bonding orbitals are generally not drawn. Looking a the MOs constucted the only ones relevant in looking at aromaticity are 17, 20, 21 and 22. Looking at t17, (totally bonding) the orbitals lie along the top and bottom of the molecule, allowing delocalisation over the whole structure. As this is a property of aromaticity this shows how MOs show the concept.
Boratabenzene
The molecule was first optimised using a high basis set and further constraints listed below, DOI:10042/21976
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -219.021 a.u. |
| RMS Gradient Norm | 0.00015769 a.u. |
| Dipole Moment | 2.85D |
| Point Group | C1 |
| Run Time | 3 minutes 15.4 seconds |
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000069 0.000300 YES
Maximum Displacement 0.000911 0.001800 YES
RMS Displacement 0.000329 0.001200 YES
Predicted change in Energy=-6.571678D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3989 -DE/DX = 0.0 !
! R2 R(1,5) 1.4053 -DE/DX = -0.0001 !
! R3 R(1,6) 1.0968 -DE/DX = 0.0001 !
! R4 R(2,7) 1.097 -DE/DX = -0.0001 !
! R5 R(2,12) 1.5137 -DE/DX = 0.0002 !
! R6 R(3,4) 1.3989 -DE/DX = 0.0 !
! R7 R(3,9) 1.097 -DE/DX = -0.0001 !
! R8 R(3,12) 1.5138 -DE/DX = 0.0001 !
! R9 R(4,5) 1.4053 -DE/DX = -0.0001 !
! R10 R(4,10) 1.0968 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0916 -DE/DX = -0.0001 !
! R12 R(8,12) 1.2185 -DE/DX = 0.0 !
! A1 A(2,1,5) 122.1394 -DE/DX = 0.0001 !
! A2 A(2,1,6) 120.4237 -DE/DX = -0.0002 !
! A3 A(5,1,6) 117.437 -DE/DX = 0.0 !
! A4 A(1,2,7) 115.9496 -DE/DX = 0.0001 !
! A5 A(1,2,12) 120.0811 -DE/DX = -0.0001 !
! A6 A(7,2,12) 123.9693 -DE/DX = -0.0001 !
! A7 A(4,3,9) 115.9541 -DE/DX = 0.0001 !
! A8 A(4,3,12) 120.0808 -DE/DX = -0.0001 !
! A9 A(9,3,12) 123.9651 -DE/DX = -0.0001 !
! A10 A(3,4,5) 122.1382 -DE/DX = 0.0001 !
! A11 A(3,4,10) 120.4266 -DE/DX = -0.0002 !
! A12 A(5,4,10) 117.4352 -DE/DX = 0.0 !
! A13 A(1,5,4) 120.4509 -DE/DX = -0.0001 !
! A14 A(1,5,11) 119.7757 -DE/DX = 0.0001 !
! A15 A(4,5,11) 119.7734 -DE/DX = 0.0001 !
! A16 A(2,12,3) 115.1096 -DE/DX = 0.0 !
! A17 A(2,12,8) 122.449 -DE/DX = 0.0 !
! A18 A(3,12,8) 122.4414 -DE/DX = 0.0 !
! D1 D(5,1,2,7) -180.0047 -DE/DX = 0.0 !
! D2 D(5,1,2,12) -0.003 -DE/DX = 0.0 !
! D3 D(6,1,2,7) 0.0006 -DE/DX = 0.0 !
! D4 D(6,1,2,12) 180.0023 -DE/DX = 0.0 !
! D5 D(2,1,5,4) 0.0096 -DE/DX = 0.0 !
! D6 D(2,1,5,11) 180.0051 -DE/DX = 0.0 !
! D7 D(6,1,5,4) 180.0044 -DE/DX = 0.0 !
! D8 D(6,1,5,11) -0.0001 -DE/DX = 0.0 !
! D9 D(1,2,12,3) -0.0017 -DE/DX = 0.0 !
! D10 D(1,2,12,8) -180.0001 -DE/DX = 0.0 !
! D11 D(7,2,12,3) 180.0001 -DE/DX = 0.0 !
! D12 D(7,2,12,8) 0.0017 -DE/DX = 0.0 !
! D13 D(9,3,4,5) 180.0078 -DE/DX = 0.0 !
! D14 D(9,3,4,10) -0.0017 -DE/DX = 0.0 !
! D15 D(12,3,4,5) 0.0064 -DE/DX = 0.0 !
! D16 D(12,3,4,10) -180.003 -DE/DX = 0.0 !
! D17 D(4,3,12,2) 0.0001 -DE/DX = 0.0 !
! D18 D(4,3,12,8) 179.9985 -DE/DX = 0.0 !
! D19 D(9,3,12,2) -180.0014 -DE/DX = 0.0 !
! D20 D(9,3,12,8) -0.003 -DE/DX = 0.0 !
! D21 D(3,4,5,1) -0.0114 -DE/DX = 0.0 !
! D22 D(3,4,5,11) -180.0069 -DE/DX = 0.0 !
! D23 D(10,4,5,1) -180.0022 -DE/DX = 0.0 !
! D24 D(10,4,5,11) 0.0023 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
To check optimisation completed further vibrational analysis was also carried out on the HPC scanner. This was also helpful as it allows us to locate the peaks expected on an IR spectra without running one and also determining the intensity of these peaks. DOI:10042/21865
Item Value Threshold Converged?
Maximum Force 0.000437 0.000450 YES
RMS Force 0.000156 0.000300 YES
Maximum Displacement 0.000855 0.001800 YES
RMS Displacement 0.000387 0.001200 YES
Predicted change in Energy=-7.057381D-07
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies --- -6.8496 0.0006 0.0007 0.0007 3.2247 5.6419 Low frequencies --- 371.2623 404.4982 565.1635
1 2 3
A A A
Frequencies -- 371.2623 404.4981 565.1635
Shown are the frequencies and their intensities alongside the IR spectra.
| No. | Frequency | Intensity | No. | Frequency | Intensity | No. | Frequency | Intensity |
|---|---|---|---|---|---|---|---|---|
| 1 | 375 | 2 | 11 | 900 | 1 | 21 | 1400 | 9 |
| 2 | 400 | 0 | 12 | 1000 | 0 | 22 | 1500 | 14 |
| 3 | 575 | 0 | 13 | 1000 | 0 | 23 | 1600 | 7 |
| 4 | 575 | 0 | 14 | 1000 | 2 | 24 | 1600 | 40 |
| 5 | 600 | 11 | 15 | 1000 | 3 | 25 | 2000 | 468 |
| 6 | 700 | 3 | 16 | 1100 | 3 | 26 | 3000 | 109 |
| 7 | 775 | 7 | 17 | 1200 | 1 | 27 | 3000 | 109 |
| 8 | 825 | 0 | 18 | 1200 | 1 | 28 | 3100 | 378 |
| 9 | 875 | 28 | 19 | 1200 | 1 | 29 | 3000 | 8 |
| 30 | 3100 | 113 |
Results from the opimisation can be seen showing the optimal bond lengths in boratabenzene,
| Bond | Bond length |
|---|---|
| B-C | 1.51A |
| C-C | 1.40A |
These bond lengths are as to be expected due to conjugation along the framework. The optimal bond angle in the ring, is 115.11° (C-B-C) The other angles inside the ring show slight differences due to the introduction of the boron atom, and the smaller angle which is now present.
Having confirmed the molecule fully optimised and having looked at the vibrational analysis of this molecule MO analysis could be done, DOI:10042/22001 . The MO'S have been tabelised below with next to their corresponding orbital number. The first 6 MO'S have been neglected as these are core MO'S and therefore do not can be discounted.
| 7 |  |
8 | 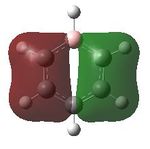 |
9 | 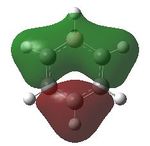 |
10 | 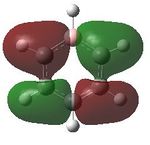 |
11 | 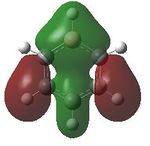 |
12 | 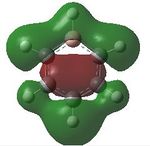
|
| 13 |  |
14 | 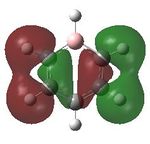 |
15 | 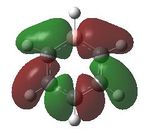 |
16 | 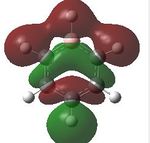 |
17 | 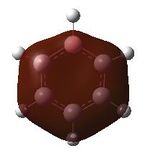 |
18 | 
|
| 19 |  |
20 |  |
21 |  |
22 | 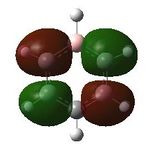
|
In order to look closely at the bonds in boratabenzene NBO analysis was carried out, this allowed us to look at the hybridization of the carbons in the atom and to see the percentage population of each atom in bonds.
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98270) BD ( 1) C 1 - C 2
( 50.77%) 0.7125* C 1 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 0.0575
0.0311 0.7869 0.0164 0.0000 0.0000
0.0020 0.0000 0.0000 -0.0150 -0.0098
( 49.23%) 0.7017* C 2 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 -0.0027
0.0269 -0.8202 -0.0353 0.0000 0.0000
-0.0006 0.0000 0.0000 -0.0173 -0.0123
2. (1.76862) BD ( 2) C 1 - C 2
( 48.13%) 0.6937* C 1 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9996 -0.0213
0.0000 -0.0031 0.0171 0.0000 0.0000
( 51.87%) 0.7202* C 2 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9998 -0.0054
0.0000 -0.0016 -0.0185 0.0000 0.0000
3. (1.97971) BD ( 1) C 1 - C 5
( 49.96%) 0.7068* C 1 s( 35.51%)p 1.82( 64.45%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 -0.6875
-0.0034 -0.4133 -0.0325 0.0000 0.0000
0.0146 0.0000 0.0000 0.0081 -0.0107
( 50.04%) 0.7074* C 5 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 0.7063
0.0327 0.3752 -0.0141 0.0000 0.0000
0.0137 0.0000 0.0000 0.0078 -0.0107
4. (1.98570) BD ( 1) C 1 - H 6
( 59.32%) 0.7702* C 1 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
-0.0003 0.5182 0.0133 -0.0012 0.7227
-0.0089 -0.4563 0.0100 0.0000 0.0000
-0.0177 0.0000 0.0000 0.0069 -0.0111
( 40.68%) 0.6378* H 6 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0026 -0.0187 0.0116 0.0000
5. (1.98420) BD ( 1) C 2 - H 7
( 59.41%) 0.7708* C 2 s( 25.38%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 0.7907
-0.0003 0.3469 0.0088 0.0000 0.0000
0.0111 0.0000 0.0000 0.0150 -0.0119
( 40.59%) 0.6371* H 7 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 -0.0192 -0.0100 0.0000
6. (1.96997) BD ( 1) C 2 - B 12
( 66.70%) 0.8167* C 2 s( 42.04%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 -0.6482 -0.0158 -0.0012 0.6115
-0.0293 -0.4524 -0.0090 0.0000 0.0000
0.0059 0.0000 0.0000 -0.0041 0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.52%)d 0.00( 0.08%)
0.0000 -0.5779 0.0059 -0.0048 -0.7056
-0.0393 0.4071 -0.0096 0.0000 0.0000
0.0230 0.0000 0.0000 -0.0082 0.0133
7. (1.98270) BD ( 1) C 3 - C 4
( 49.23%) 0.7017* C 3 s( 32.49%)p 2.08( 67.46%)d 0.00( 0.05%)
0.0000 0.5697 -0.0200 0.0010 0.0025
-0.0269 -0.8201 -0.0353 0.0000 0.0000
0.0006 0.0000 0.0000 -0.0173 -0.0123
( 50.77%) 0.7125* C 4 s( 37.60%)p 1.66( 62.37%)d 0.00( 0.03%)
-0.0001 0.6131 -0.0079 0.0007 -0.0573
-0.0311 0.7869 0.0164 0.0000 0.0000
-0.0020 0.0000 0.0000 -0.0150 -0.0098
8. (1.76868) BD ( 2) C 3 - C 4
( 51.87%) 0.7202* C 3 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 -0.0001 0.0000 0.9998 -0.0054
0.0000 0.0016 -0.0185 0.0000 0.0000
( 48.13%) 0.6938* C 4 s( 0.00%)p 1.00( 99.97%)d 0.00( 0.03%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9996 -0.0213
0.0000 0.0031 0.0171 0.0000 0.0000
9. (1.98420) BD ( 1) C 3 - H 9
( 59.41%) 0.7708* C 3 s( 25.39%)p 2.94( 74.57%)d 0.00( 0.05%)
-0.0003 0.5038 -0.0051 -0.0025 -0.7906
0.0003 0.3472 0.0088 0.0000 0.0000
-0.0111 0.0000 0.0000 0.0149 -0.0119
( 40.59%) 0.6371* H 9 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0005 0.0192 -0.0100 0.0000
10. (1.96996) BD ( 1) C 3 - B 12
( 66.70%) 0.8167* C 3 s( 42.03%)p 1.38( 57.96%)d 0.00( 0.01%)
0.0000 0.6481 0.0158 0.0012 0.6117
-0.0293 0.4522 0.0090 0.0000 0.0000
0.0059 0.0000 0.0000 0.0041 -0.0057
( 33.30%) 0.5771* B 12 s( 33.40%)p 1.99( 66.53%)d 0.00( 0.08%)
0.0000 0.5779 -0.0059 0.0048 -0.7057
-0.0393 -0.4070 0.0096 0.0000 0.0000
0.0230 0.0000 0.0000 0.0082 -0.0133
11. (1.97970) BD ( 1) C 4 - C 5
( 49.96%) 0.7068* C 4 s( 35.50%)p 1.82( 64.46%)d 0.00( 0.04%)
-0.0001 0.5958 -0.0075 0.0006 0.6874
0.0034 -0.4135 -0.0325 0.0000 0.0000
-0.0146 0.0000 0.0000 0.0081 -0.0107
( 50.04%) 0.7074* C 5 s( 35.88%)p 1.79( 64.09%)d 0.00( 0.04%)
-0.0001 0.5989 -0.0072 0.0010 -0.7062
-0.0327 0.3754 -0.0141 -0.0001 0.0000
-0.0137 0.0000 0.0000 0.0078 -0.0107
12. (1.98570) BD ( 1) C 4 - H 10
( 59.32%) 0.7702* C 4 s( 26.88%)p 2.72( 73.08%)d 0.00( 0.05%)
0.0003 -0.5182 -0.0133 0.0012 0.7228
-0.0089 0.4562 -0.0100 0.0000 0.0000
-0.0177 0.0000 0.0000 -0.0069 0.0111
( 40.68%) 0.6378* H 10 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0026 -0.0187 -0.0116 0.0000
13. (1.98507) BD ( 1) C 5 - H 11
( 59.44%) 0.7710* C 5 s( 28.22%)p 2.54( 71.74%)d 0.00( 0.04%)
0.0004 -0.5311 -0.0116 0.0020 0.0001
0.0000 0.8469 -0.0076 0.0000 0.0000
0.0000 0.0000 0.0000 0.0178 0.0110
( 40.56%) 0.6369* H 11 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0011 0.0000 -0.0217 0.0000
14. (1.98604) BD ( 1) H 8 - B 12
( 55.09%) 0.7422* H 8 s( 99.97%)p 0.00( 0.03%)
0.9998 0.0001 0.0000 -0.0180 0.0000
( 44.91%) 0.6702* B 12 s( 33.16%)p 2.01( 66.78%)d 0.00( 0.06%)
-0.0005 0.5758 0.0069 -0.0060 0.0001
0.0000 0.8172 -0.0016 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0213 -0.0105
As can be seen from the data above, the addition of a B atom has changed the values of sp character and also the percentage of each carbon atom in the bond. Unlike in benzene the carbon bond does not contain an exact 50% separation of each carbon atom, although the value has not shifted significantly there is now not an equal split in the bond property, this could be affected by the nature of the Boron atom now in the ring. Looking at the hybiridisation of the atoms in this molecule it seems that instead of 3 double bonds it now appears to have only two, therefore giving some of the Carbons an sp2 hybiridised structure, shown by the 26:78 ratio of sp character. To look at the charge distribution in the Boratabenzene molecule an image was created showing the different charges by corresponding colours, red for -0.588D and green for 0.588D, this can be seen on the right with the actual values tabulated below.
| Atom | Charge |
|---|---|
| Boron | 0.20D |
| Carbon ortho to Boron | -0.59D |
| Carbon meta to Boron | -0.25D |
| Carbon para to Boron | -0.34D |
| Hydrogen on Boron | -0.1D |
| Hydrogen on ortho Carbon | 0.18D |
| Hydrogen on meta Carbon | 0.18D |
| Hydrogen on para Carbon | 0.19D |
As can be seen the charge distribution is now no longer equal, due to the introduction of the electropositive boron, this can be seen clearly as carbons closer to the boron have an increased negative charge than those further around the framework.
Pyridinium
This molecule was produced in Gaussview via a benzene ring with one carbon swapped for a Nitrogen atom, due to this change in atoms it was necessary to ensure the molecule was given a charge of +1, ensuring the charge was present in the method tab of all scans. The molecule was optimised under a high basis set of 6-31G(d,p) through the HPC scanner with the following constraints, more detail can be found here DOI:10042/21996
| File Type | .log |
| Calculation Type | FOPT |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -248.668 a.u. |
| RMS Gradient Norm | 0.00003914 a.u. |
| Dipole Moment | 1.87D |
| Point Group | C1 |
| Run Time | 4 minutes 3 seconds |
Item Value Threshold Converged?
Maximum Force 0.000064 0.000450 YES
RMS Force 0.000023 0.000300 YES
Maximum Displacement 0.000702 0.001800 YES
RMS Displacement 0.000176 0.001200 YES
Predicted change in Energy=-6.970286D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3839 -DE/DX = 0.0 !
! R2 R(1,5) 1.3988 -DE/DX = 0.0 !
! R3 R(1,6) 1.0835 -DE/DX = 0.0 !
! R4 R(2,7) 1.0832 -DE/DX = 0.0 !
! R5 R(2,12) 1.3523 -DE/DX = 0.0001 !
! R6 R(3,4) 1.3838 -DE/DX = 0.0 !
! R7 R(3,9) 1.0832 -DE/DX = 0.0 !
! R8 R(3,12) 1.3524 -DE/DX = 0.0 !
! R9 R(4,5) 1.3988 -DE/DX = 0.0 !
! R10 R(4,10) 1.0835 -DE/DX = 0.0 !
! R11 R(5,11) 1.0852 -DE/DX = 0.0 !
! R12 R(8,12) 1.0169 -DE/DX = 0.0 !
! A1 A(2,1,5) 119.082 -DE/DX = 0.0 !
! A2 A(2,1,6) 119.4193 -DE/DX = 0.0001 !
! A3 A(5,1,6) 121.4987 -DE/DX = -0.0001 !
! A4 A(1,2,7) 123.9293 -DE/DX = 0.0 !
! A5 A(1,2,12) 119.2363 -DE/DX = 0.0 !
! A6 A(7,2,12) 116.8344 -DE/DX = 0.0 !
! A7 A(4,3,9) 123.9326 -DE/DX = 0.0 !
! A8 A(4,3,12) 119.2354 -DE/DX = 0.0 !
! A9 A(9,3,12) 116.8319 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.0827 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.4215 -DE/DX = 0.0 !
! A12 A(5,4,10) 121.4958 -DE/DX = -0.0001 !
! A13 A(1,5,4) 120.0549 -DE/DX = 0.0 !
! A14 A(1,5,11) 119.9741 -DE/DX = 0.0 !
! A15 A(4,5,11) 119.971 -DE/DX = 0.0 !
! A16 A(2,12,3) 123.3087 -DE/DX = 0.0 !
! A17 A(2,12,8) 118.3464 -DE/DX = 0.0 !
! A18 A(3,12,8) 118.3449 -DE/DX = 0.0 !
! D1 D(5,1,2,7) -180.0012 -DE/DX = 0.0 !
! D2 D(5,1,2,12) 0.0031 -DE/DX = 0.0 !
! D3 D(6,1,2,7) -0.0013 -DE/DX = 0.0 !
! D4 D(6,1,2,12) 180.003 -DE/DX = 0.0 !
! D5 D(2,1,5,4) -0.0004 -DE/DX = 0.0 !
! D6 D(2,1,5,11) -180.0014 -DE/DX = 0.0 !
! D7 D(6,1,5,4) -180.0003 -DE/DX = 0.0 !
! D8 D(6,1,5,11) -0.0013 -DE/DX = 0.0 !
! D9 D(1,2,12,3) -0.0045 -DE/DX = 0.0 !
! D10 D(1,2,12,8) -180.0032 -DE/DX = 0.0 !
! D11 D(7,2,12,3) -180.0005 -DE/DX = 0.0 !
! D12 D(7,2,12,8) 0.0008 -DE/DX = 0.0 !
! D13 D(9,3,4,5) 180.0009 -DE/DX = 0.0 !
! D14 D(9,3,4,10) -0.0003 -DE/DX = 0.0 !
! D15 D(12,3,4,5) 0.0 -DE/DX = 0.0 !
! D16 D(12,3,4,10) -180.0012 -DE/DX = 0.0 !
! D17 D(4,3,12,2) 0.0029 -DE/DX = 0.0 !
! D18 D(4,3,12,8) 180.0016 -DE/DX = 0.0 !
! D19 D(9,3,12,2) 180.002 -DE/DX = 0.0 !
! D20 D(9,3,12,8) 0.0008 -DE/DX = 0.0 !
! D21 D(3,4,5,1) -0.0011 -DE/DX = 0.0 !
! D22 D(3,4,5,11) -180.0001 -DE/DX = 0.0 !
! D23 D(10,4,5,1) 180.0001 -DE/DX = 0.0 !
! D24 D(10,4,5,11) 0.0011 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency analysis was then undertaken on the molecule using the same constraints as discussed earlier but the full file can be found here DOI:10042/21997
Item Value Threshold Converged?
Maximum Force 0.000129 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.000791 0.001800 YES
RMS Displacement 0.000229 0.001200 YES
Predicted change in Energy=-6.662774D-08
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies --- -9.4949 -5.2553 -0.0012 -0.0008 0.0000 3.9984 Low frequencies --- 391.9053 404.3504 620.1996
1 2 3
A A A
Frequencies -- 391.9053 404.3504 620.1996
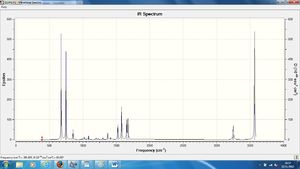
A summary of the vibrations in the pyridinium molecule and their corresponding intensities, also the IR spectra.
| No. | Frequency | Intensity | No. | Frequency | Intensity | No. | Frequency | Intensity |
|---|---|---|---|---|---|---|---|---|
| 1 | 400 | 1 | 11 | 1000 | 4 | 21 | 1500 | 21 |
| 2 | 400 | 0 | 12 | 1000 | 0 | 22 | 1600 | 48 |
| 3 | 625 | 0 | 13 | 1100 | 0 | 23 | 1700 | 32 |
| 4 | 650 | 0 | 14 | 1100 | 3 | 24 | 1700 | 34 |
| 5 | 700 | 90 | 15 | 1100 | 4 | 25 | 3200 | 0 |
| 6 | 750 | 82 | 16 | 1200 | 3 | 26 | 3200 | 1 |
| 7 | 875 | 11 | 17 | 1200 | 2 | 27 | 3200 | 11 |
| 8 | 900 | 0 | 18 | 1300 | 3 | 28 | 3300 | 20 |
| 9 | 1000 | 2 | 19 | 1400 | 11 | 29 | 3300 | 0 |
| 30 | 3600 | 158 |
The table of frequencies above shows the number of expected peaks seen on the IR spectra also shown above. There is an obvious difference in this table compared to that of benzene this is the larger number of peaks seen. The reason for this is the different environments of the hydrogens caused by the ortho/meta/para positions the carbons are now in to the Nitrogen atom.
From all data above the optimal bond lengths in this molecule are as follows,
| Bond | Bond length |
|---|---|
| N-C | 1.35A |
| C-C | 1.40A |
The C-C bond lengths are as expected, conjugate system. The shorter C-N bond length is due to the electronegative nature of the nitrogen atom. With the optimal C-N-C bond angle being 122.5°. The C-C-C bond angles are slightly different throughout the chain due to the N atom being included.
MO analysis was then undertaken on the HPC scanner using the same set up as for previous scans, full details of files can be found here DOI:10042/22014 . Shown below are the relevant occupied MO's for pyridinium, and the LUMO, the first 6 have not been included as these are the core orbitals and are not involved in the bonding.
| 7 | 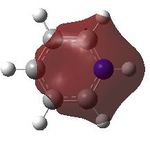 |
8 |  |
9 | 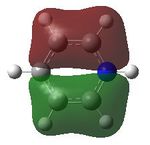 |
10 | 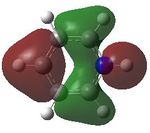 |
11 | 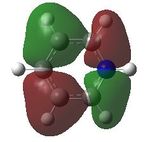 |
12 | 
|
| 13 |  |
14 | 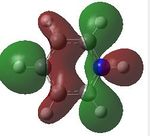 |
15 | 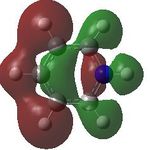 |
16 | 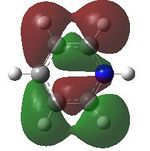 |
17 | 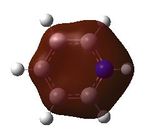 |
18 | 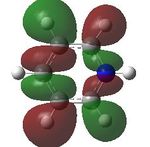
|
| 19 | 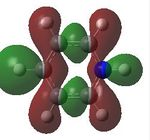 |
20 | 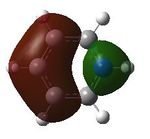 |
21 |  |
22 | 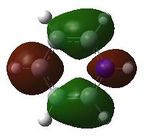
|
Natural Bond Analysis was then carried out on the pyridinium molecule with focus being on the population of each atom in bonds and of the hybridisation in the molecule. A brief summary of the results can be seen below
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H6N)
1. BD ( 1) C 1 - C 2 1.98297 -0.92648 120(v),119(v),108(g),63(v)
110(g),97(v),109(g),62(v)
111(g),96(v)
2. BD ( 2) C 1 - C 2 1.61439 -0.46666 117(v),115(v),99(v),107(g)
67(v)
3. BD ( 1) C 1 - C 5 1.98249 -0.90379 110(v),118(v),106(g),33(v)
116(g),109(g),53(v),52(v)
119(g)
4. BD ( 1) C 1 - H 6 1.97822 -0.71855 111(v),116(v),62(v),32(v)
108(g),106(g)
5. BD ( 1) C 2 - H 7 1.98154 -0.75115 114(v),108(v),22(v),106(g)
96(v)
6. BD ( 1) C 2 - N 12 1.98861 -1.06562 114(g),42(v),109(v),23(v)
113(v),44(v),106(g),120(g)
7. BD ( 1) C 3 - C 4 1.98297 -0.92650 120(v),119(v),116(g),63(v)
113(g),97(v),118(g),62(v)
114(g),96(v)
8. BD ( 1) C 3 - H 9 1.98154 -0.75115 111(v),116(v),52(v),112(g)
96(v)
9. BD ( 1) C 3 - N 12 1.98861 -1.06559 111(g),32(v),118(v),53(v)
110(v),33(v),112(g),120(g)
10. BD ( 2) C 3 - N 12 1.82443 -0.56811 107(v),117(v),34(v),56(v)
11. BD ( 1) C 4 - C 5 1.98249 -0.90377 113(v),109(v),112(g),44(v)
108(g),118(g),23(v),22(v)
119(g)
12. BD ( 2) C 4 - C 5 1.54874 -0.44891 115(v),107(v),46(v),26(v)
13. BD ( 1) C 4 - H 10 1.97822 -0.71855 114(v),108(v),62(v),42(v)
116(g),112(g)
14. BD ( 1) C 5 - H 11 1.98141 -0.71806 106(v),112(v),22(v),52(v)
108(g),116(g)
15. BD ( 1) H 8 - N 12 1.98630 -0.89230 106(v),112(v),32(v),42(v)
16. CR ( 1) C 1 1.99913 -10.26482 63(v),33(v),32(v),110(v)
116(v),72(v),119(v),111(v)
17. CR ( 1) C 2 1.99918 -10.32333 23(v),114(v),120(v),108(v)
106(g),97(v),76(v),109(v)
18. CR ( 1) C 3 1.99918 -10.32332 53(v),111(v),120(v),116(v)
112(g),97(v),84(v),118(v)
19. CR ( 1) C 4 1.99913 -10.26482 63(v),44(v),42(v),113(v)
108(v),88(v),119(v),114(v)
20. CR ( 1) C 5 1.99914 -10.27397 23(v),53(v),106(v),112(v)
109(v),118(v),92(v)
21. CR ( 1) N 12 1.99937 -14.46220 33(v),44(v)
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98297) BD ( 1) C 1 - C 2
( 49.58%) 0.7042* C 1 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 0.8145
0.0194 0.0007 -0.0320 0.0000 0.0000
0.0047 0.0000 0.0000 0.0179 -0.0119
( 50.42%) 0.7100* C 2 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 -0.7832
-0.0046 -0.0140 -0.0331 0.0000 0.0000
-0.0053 0.0000 0.0000 0.0168 -0.0095
2. (1.61439) BD ( 2) C 1 - C 2
( 52.23%) 0.7227* C 1 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0068
0.0000 0.0191 0.0159 0.0000 0.0000
( 47.77%) 0.6912* C 2 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9995 -0.0175
0.0000 -0.0196 0.0153 0.0000 0.0000
3. (1.98249) BD ( 1) C 1 - C 5
( 50.26%) 0.7089* C 1 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 -0.4184
-0.0371 0.6898 -0.0068 0.0000 0.0000
-0.0122 0.0000 0.0000 -0.0118 -0.0115
( 49.74%) 0.7053* C 5 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 0.3934
-0.0234 -0.7064 -0.0290 0.0000 0.0000
-0.0169 0.0000 0.0000 -0.0060 -0.0113
4. (1.97822) BD ( 1) C 1 - H 6
( 64.83%) 0.8052* C 1 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
0.0003 -0.5636 -0.0138 0.0005 0.3989
-0.0072 0.7228 -0.0181 0.0000 0.0000
-0.0109 0.0000 0.0000 0.0085 0.0099
( 35.17%) 0.5930* H 6 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0016 -0.0116 -0.0208 0.0000
5. (1.98154) BD ( 1) C 2 - H 7
( 64.26%) 0.8016* C 2 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
0.0004 -0.5780 -0.0180 0.0017 -0.4693
0.0193 0.6666 -0.0183 0.0000 0.0000
0.0164 0.0000 0.0000 0.0019 0.0092
( 35.74%) 0.5978* H 7 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 0.0128 -0.0209 0.0000
6. (1.98861) BD ( 1) C 2 - N 12
( 36.68%) 0.6057* C 2 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5293 -0.0335 -0.0013 0.4043
0.0563 0.7416 0.0277 0.0000 0.0000
0.0252 0.0000 0.0000 -0.0185 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6047 -0.0037 0.0006 -0.3660
0.0187 -0.7069 -0.0132 0.0000 0.0000
0.0107 0.0000 0.0000 -0.0059 -0.0115
7. (1.98297) BD ( 1) C 3 - C 4
( 50.42%) 0.7100* C 3 s( 38.49%)p 1.60( 61.47%)d 0.00( 0.04%)
-0.0001 0.6204 -0.0023 0.0030 -0.7832
-0.0046 0.0142 0.0331 0.0000 0.0000
0.0053 0.0000 0.0000 0.0168 -0.0095
( 49.58%) 0.7042* C 4 s( 33.47%)p 1.99( 66.48%)d 0.00( 0.05%)
0.0000 0.5784 -0.0119 -0.0002 0.8145
0.0194 -0.0009 0.0320 0.0000 0.0000
-0.0047 0.0000 0.0000 0.0179 -0.0119
8. (1.98154) BD ( 1) C 3 - H 9
( 64.26%) 0.8016* C 3 s( 33.44%)p 1.99( 66.52%)d 0.00( 0.04%)
-0.0004 0.5780 0.0180 -0.0017 0.4694
-0.0193 0.6664 -0.0183 0.0000 0.0000
0.0164 0.0000 0.0000 -0.0019 -0.0092
( 35.74%) 0.5978* H 9 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0018 -0.0128 -0.0209 0.0000
9. (1.98861) BD ( 1) C 3 - N 12
( 36.68%) 0.6057* C 3 s( 28.13%)p 2.55( 71.74%)d 0.00( 0.13%)
-0.0001 0.5293 -0.0335 -0.0013 0.4042
0.0563 -0.7417 -0.0277 0.0000 0.0000
-0.0251 0.0000 0.0000 -0.0185 -0.0179
( 63.32%) 0.7957* N 12 s( 36.56%)p 1.73( 63.41%)d 0.00( 0.03%)
-0.0001 0.6046 -0.0037 0.0006 -0.3658
0.0187 0.7069 0.0132 0.0000 0.0000
-0.0107 0.0000 0.0000 -0.0059 -0.0115
10. (1.82443) BD ( 2) C 3 - N 12
( 28.55%) 0.5343* C 3 s( 0.00%)p 1.00( 99.83%)d 0.00( 0.17%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9991 0.0132
0.0000 0.0102 -0.0394 0.0000 0.0000
( 71.45%) 0.8453* N 12 s( 0.00%)p 1.00( 99.98%)d 0.00( 0.02%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9999 0.0036
0.0000 -0.0128 0.0077 0.0000 0.0000
11. (1.98249) BD ( 1) C 4 - C 5
( 50.26%) 0.7089* C 4 s( 34.73%)p 1.88( 65.23%)d 0.00( 0.04%)
0.0000 0.5893 -0.0066 0.0009 -0.4185
-0.0371 -0.6897 0.0068 0.0000 0.0000
0.0122 0.0000 0.0000 -0.0118 -0.0115
( 49.74%) 0.7053* C 5 s( 34.45%)p 1.90( 65.51%)d 0.00( 0.04%)
0.0000 0.5869 -0.0086 0.0005 0.3935
-0.0234 0.7063 0.0290 0.0000 0.0000
0.0169 0.0000 0.0000 -0.0060 -0.0113
12. (1.54874) BD ( 2) C 4 - C 5
( 54.27%) 0.7367* C 4 s( 0.00%)p 1.00( 99.94%)d 0.00( 0.06%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0080
0.0000 0.0086 -0.0228 0.0000 0.0000
( 45.73%) 0.6762* C 5 s( 0.00%)p 1.00( 99.93%)d 0.00( 0.07%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.9997 -0.0036
0.0000 0.0241 0.0100 0.0000 0.0000
13. (1.97822) BD ( 1) C 4 - H 10
( 64.83%) 0.8052* C 4 s( 31.78%)p 2.15( 68.19%)d 0.00( 0.03%)
-0.0003 0.5636 0.0138 -0.0005 -0.3987
0.0072 0.7229 -0.0181 0.0000 0.0000
-0.0109 0.0000 0.0000 -0.0085 -0.0099
( 35.17%) 0.5930* H 10 s( 99.94%)p 0.00( 0.06%)
0.9997 0.0016 0.0116 -0.0208 0.0000
14. (1.98141) BD ( 1) C 5 - H 11
( 64.64%) 0.8040* C 5 s( 31.07%)p 2.22( 68.89%)d 0.00( 0.03%)
0.0003 -0.5573 -0.0131 0.0007 0.8298
-0.0198 -0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 -0.0153 0.0101
( 35.36%) 0.5947* H 11 s( 99.94%)p 0.00( 0.06%)
-0.9997 -0.0018 -0.0242 0.0000 0.0000
15. (1.98630) BD ( 1) H 8 - N 12
( 25.41%) 0.5041* H 8 s( 99.88%)p 0.00( 0.12%)
0.9994 -0.0064 -0.0342 0.0000 0.0000
( 74.59%) 0.8637* N 12 s( 26.82%)p 2.73( 73.16%)d 0.00( 0.02%)
-0.0002 0.5178 0.0066 -0.0013 0.8553
-0.0091 -0.0001 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0115 -0.0106
From the data above is can be seen that pyridinium seems to be more similar in structure to benzene than boratabenzene, seen through its larger sp2 hybridisation similarities. NBO is also used to look at the charge distribution in the molecule, giving individual values for charges of molecules, however it can be easier to see the general trend of charge with a colour key as shown, red = -0.483D and green = 0.483D.
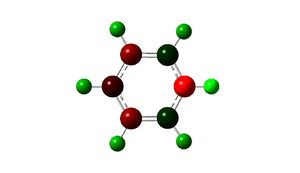
| Atom | Charge |
|---|---|
| Nitrogen | -0.48D |
| Carbon ortho to Nitrogen | 0.07D |
| Carbon meta to Nitrogen | -0.24D |
| Carbon para to Nitrogen | -0.12D |
| Hydrogen on Nitrogen | 0.48D |
| Hydrogen on ortho Carbon | 0.29D |
| Hydrogen on meta Carbon | 0.30D |
| Hydrogen on para Carbon | 0.29D |
The charge distribution in the molecule is again not equal, due to the introduction of the electronegative nitrogen, this has an affect on the chain causing the carbons closest to it to exhibit more positive charges than those on the edge of the framework furthest from the nitrogen. The hydrogen bound to the nitrogen has the highest positive charge character due to the electronegative nature of the nitrogen.
Borazine
A molecule of Borazine was created in Gaussview using a template for benzene and switching the carbon atoms for the respective boron and nitrogen. It was optimised using a high basis set of 6-31G(d,p) on the HPC scanner.
| File Type | .log |
| Calculation Type | FREQ |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Energy | -242.685 a.u. |
| RMS Gradient Norm | 0.00006608 a.u. |
| Dipole Moment | 0D |
| Point Group | |
| Run Time | 5 minutes 58 seconds |
Item Value Threshold Converged?
Maximum Force 0.000093 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.000341 0.001800 YES
RMS Displacement 0.000096 0.001200 YES
Predicted change in Energy=-1.028626D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,8) 1.1949 -DE/DX = 0.0001 !
! R2 R(2,12) 1.0097 -DE/DX = 0.0 !
! R3 R(3,9) 1.1949 -DE/DX = 0.0001 !
! R4 R(4,11) 1.0097 -DE/DX = 0.0 !
! R5 R(5,7) 1.1949 -DE/DX = 0.0001 !
! R6 R(6,10) 1.0097 -DE/DX = 0.0 !
! R7 R(7,10) 1.4307 -DE/DX = -0.0001 !
! R8 R(7,11) 1.4306 -DE/DX = -0.0001 !
! R9 R(8,10) 1.4306 -DE/DX = 0.0 !
! R10 R(8,12) 1.4307 -DE/DX = 0.0 !
! R11 R(9,11) 1.4307 -DE/DX = -0.0001 !
! R12 R(9,12) 1.4307 -DE/DX = 0.0 !
! A1 A(5,7,10) 121.4373 -DE/DX = 0.0 !
! A2 A(5,7,11) 121.4436 -DE/DX = 0.0 !
! A3 A(10,7,11) 117.119 -DE/DX = 0.0001 !
! A4 A(1,8,10) 121.4414 -DE/DX = 0.0 !
! A5 A(1,8,12) 121.4399 -DE/DX = 0.0 !
! A6 A(10,8,12) 117.1187 -DE/DX = 0.0 !
! A7 A(3,9,11) 121.4337 -DE/DX = 0.0 !
! A8 A(3,9,12) 121.4348 -DE/DX = 0.0 !
! A9 A(11,9,12) 117.1314 -DE/DX = 0.0 !
! A10 A(6,10,7) 118.5563 -DE/DX = 0.0 !
! A11 A(6,10,8) 118.5584 -DE/DX = 0.0 !
! A12 A(7,10,8) 122.8853 -DE/DX = 0.0 !
! A13 A(4,11,7) 118.567 -DE/DX = 0.0 !
! A14 A(4,11,9) 118.5613 -DE/DX = 0.0 !
! A15 A(7,11,9) 122.8717 -DE/DX = 0.0 !
! A16 A(2,12,8) 118.5622 -DE/DX = 0.0 !
! A17 A(2,12,9) 118.5639 -DE/DX = 0.0 !
! A18 A(8,12,9) 122.8739 -DE/DX = 0.0 !
! D1 D(5,7,10,6) 0.0003 -DE/DX = 0.0 !
! D2 D(5,7,10,8) 179.9967 -DE/DX = 0.0 !
! D3 D(11,7,10,6) -179.9996 -DE/DX = 0.0 !
! D4 D(11,7,10,8) -0.0033 -DE/DX = 0.0 !
! D5 D(5,7,11,4) 0.0043 -DE/DX = 0.0 !
! D6 D(5,7,11,9) -179.9997 -DE/DX = 0.0 !
! D7 D(10,7,11,4) -179.9957 -DE/DX = 0.0 !
! D8 D(10,7,11,9) 0.0002 -DE/DX = 0.0 !
! D9 D(1,8,10,6) -0.0025 -DE/DX = 0.0 !
! D10 D(1,8,10,7) -179.9989 -DE/DX = 0.0 !
! D11 D(12,8,10,6) 179.9973 -DE/DX = 0.0 !
! D12 D(12,8,10,7) 0.0009 -DE/DX = 0.0 !
! D13 D(1,8,12,2) -0.0061 -DE/DX = 0.0 !
! D14 D(1,8,12,9) -179.9955 -DE/DX = 0.0 !
! D15 D(10,8,12,2) 179.9942 -DE/DX = 0.0 !
! D16 D(10,8,12,9) 0.0048 -DE/DX = 0.0 !
! D17 D(3,9,11,4) 0.0005 -DE/DX = 0.0 !
! D18 D(3,9,11,7) -179.9954 -DE/DX = 0.0 !
! D19 D(12,9,11,4) -179.9991 -DE/DX = 0.0 !
! D20 D(12,9,11,7) 0.005 -DE/DX = 0.0 !
! D21 D(3,9,12,2) 0.0034 -DE/DX = 0.0 !
! D22 D(3,9,12,8) 179.9928 -DE/DX = 0.0 !
! D23 D(11,9,12,2) -179.997 -DE/DX = 0.0 !
! D24 D(11,9,12,8) -0.0075 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
From the optimisation data it can be seen that the optimal B-N bond distance is 1.431A, the optimal B-H is 1.19A, and the optimal N-H is 1.01A, and the optimal angle of the B-N-B bond is 122.87o, with the optimal angle of N-B-N being 117.12o.
The bond lengths are as expected, from literature vlaues[3] so are the bond angles, which are relatively similar to benzene, showing conjugation.
As well as optimising the molecule we looked at its vibrational data, this also helped to prove the molecule had optimised correctly.
Item Value Threshold Converged?
Maximum Force 0.000191 0.000450 YES
RMS Force 0.000063 0.000300 YES
Maximum Displacement 0.000374 0.001800 YES
RMS Displacement 0.000130 0.001200 YES
Predicted change in Energy=-1.092161D-07
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Low frequencies --- -4.9806 0.0006 0.0008 0.0013 6.7925 8.9742 Low frequencies --- 289.5802 289.6774 404.2782
1 2 3
A A A
Frequencies -- 289.5802 289.6774 404.2782
As seen from the data the calculations all ran correctly, there are no negative frequencies seen and all have the required difference of a magnitude of ten. Shown below are the frequencies present with their intensities, also shown is the IR spectra.

| No. | Frequency | Intensity | No. | Frequency | Intensity | No. | Frequency | Intensity |
|---|---|---|---|---|---|---|---|---|
| 1 | 289.58 | 0 | 11 | 927.52 | 0 | 21 | 1400.12 | 10.88 |
| 2 | 289.68 | 0 | 12 | 936.87 | 236 15 | 22 | 1400.19 | 10.93 |
| 3 | 404.28 | 23.55 | 13 | 944.49 | 0.0 | 23 | 1492.23 | 493.98 |
| 4 | 525.04 | 0.64 | 14 | 944.59 | 0 | 24 | 1492.26 | 494.02 |
| 5 | 525.11 | 0.63 | 15 | 944.95 | 0 | 25 | 2641.15 | 283.54 |
| 6 | 710.23 | 0 | 16 | 1051.85 | 0 | 26 | 2641.20 | 283.58 |
| 7 | 710.31 | 0 | 17 | 1080.62 | 0.20 | 27 | 2651.09 | 0 |
| 8 | 732.40 | 59.89 | 18 | 1080.70 | 0.20 | 28 | 3641.70 | 0.04 |
| 9 | 864.46 | 0 | 19 | 1245.33 | 0 | 29 | 3643.49 | 39.69 |
| 30 | 3643.57 | 39.72 |
MO analysis was undertaken, shown are the orbitals 7-22, leaving out the 6 core orbitals and only including one unoccupied one.
| 7 | 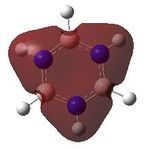 |
8 | 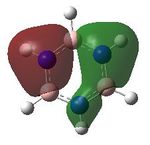 |
9 | 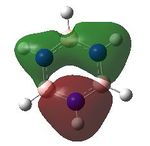 |
10 | 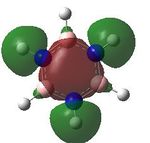 |
11 | 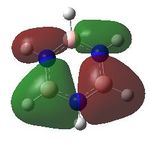 |
12 | 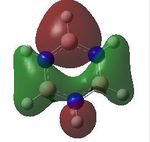
|
| 13 |  |
14 | 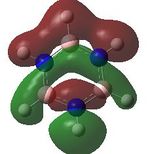 |
15 | 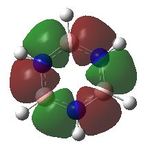 |
16 | 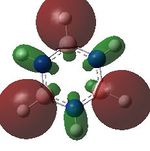 |
17 |  |
18 | 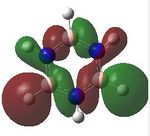
|
| 19 | 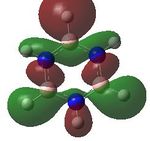 |
20 | 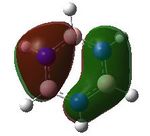 |
21 | 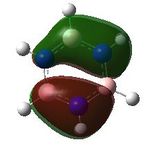 |
22 | 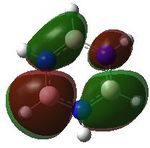
|
Looking at the NBO analysis we gained further information about the population of the molecules,
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
H 1 -0.07654 0.00000 1.07585 0.00069 1.07654
H 2 0.43198 0.00000 0.56573 0.00228 0.56802
H 3 -0.07655 0.00000 1.07586 0.00069 1.07655
H 4 0.43198 0.00000 0.56574 0.00228 0.56802
H 5 -0.07654 0.00000 1.07585 0.00069 1.07654
H 6 0.43198 0.00000 0.56574 0.00228 0.56802
B 7 0.74700 1.99917 2.23862 0.01520 4.25300
B 8 0.74695 1.99917 2.23868 0.01521 4.25305
B 9 0.74696 1.99917 2.23866 0.01521 4.25304
N 10 -1.10241 1.99943 6.09821 0.00478 8.10241
N 11 -1.10241 1.99943 6.09820 0.00478 8.10241
N 12 -1.10240 1.99943 6.09819 0.00478 8.10240
=======================================================================
* Total * 0.00000 11.99579 29.93532 0.06889 42.00000
Natural Population
--------------------------------------------------------
Core 11.99579 ( 99.9649% of 12)
Valence 29.93532 ( 99.7844% of 30)
Natural Minimal Basis 41.93111 ( 99.8360% of 42)
Natural Rydberg Basis 0.06889 ( 0.1640% of 42)
--------------------------------------------------------
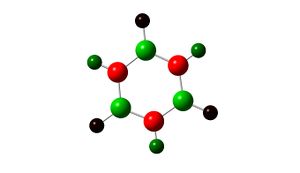
The charges on atoms in a Borazine molecule are different to those seen previously as the prescence of different atoms in the main frame of the ring affects the charges felt by the hydrogens. The charges can be seen broadly looking at the picture below which indicates the more polar ends of the molecule in different colours, red = -1.102D and green = + 1.102D.
| Atom | Charge |
|---|---|
| Boron | +0.747D |
| Hydrogen's connected to Boron | -0.077D |
| Nitrogen | -1.102D |
| Hydrogen's connected to Nitrogen | +0.432D |
The framework shows symmetric charge distribution further agreeing with the conjugation of the system, also logically it can be seen that nitrogen is electronegative, boron is electropositive and so these two should cancel each other out. The hyhdrogens bonded to the boron have a negative value (boron-electropositive) this is oppossite for the hyrdogens bonded to nitrogen.
Charge Distribution Comparison
Benzene has a symmetric charge distribution, due to its point group, conjugation of its system and aromatity. Its charge distribution is most similar to that of borazine, with borazine also having a symmetric charge distribution. Although boratbenzene's charge distribution shows similar qualities to that of benzene the appearance of a heteroatom, causes individual charges to be changed, negative closest to the boron and more positive further in the framework. Pyridinium is similar to boratabenzene in the sense the appearance of a heteroatom cause charge distribution to be varied, however this is in the opposite way to boratabenzene.
Comparing MOs
| MO | Benzene | Boratabenzene | Pyridinium | Borazine |
|---|---|---|---|---|
| 17 | 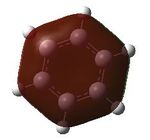 |
 |
 |

|
| 18 | 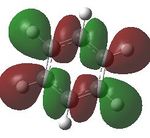 |
 |
 |

|
| 19 | 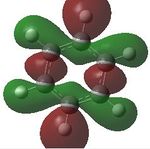 |
 |
 |

|
17- Totally bonding
This orbital is the totally bonding orbital,known from classica LCAO approach and calulation, the individual MO for each structure can be seen above. For this orbital the MOs look similar due to the nature of a totally bonding pi orbital. All p orbitals are aligned paralle to each other therefore there is not much maneuver in the picture of the orbitals. The only slight variation that can be seen is in the size of the orbitals around the heteroatoms, this is for obvious size reasons, (nitrogen is larger therefore slightly larger orbitals)
18
For this orbital the benzene and pyridinium show the most similar MO, with an almost butterfly shape of orbitals, this can be attributed to the low electron density of the hydrogen attached to the nitrogen due to its electron withdrawing effect. As opposed to the boratabenzne, which has a hydrgogen with more electron density therefore more orbital character on the MO diagram. Borazine due to the alternating nitrogen and hydrogen has properties of both pyridinium and boratabenzene, increased orbitals around the hydrgens connected to boron.
19
As with the other orbitals, the benzene and pyridinium show the moset similarities, with the only observable difference being that of the orbital over the hydrogen connected to the nitrogen, this orbital is smaller than that seen in benzene, due to the electronegative nature of nitrogen, the opposite can be seen in boratabenzene due to the electropositive nature of boron compared to carbon.
Thinking about the LCAOs of the MOs, these will change more between the molecules. In benzene the contributing atomic orbitals are are all equivalent, all coming from the s and p orbitals of carbon, and the s orbitals of hydrogen. For obvious reasons this changes for the other molecules, boratabenzene and pyridinium have the addition of a heteroatom, which has a different energy to carbon, (nitrogen - more electronegatve, lower, boron - higher). This will give a poorer overlap and therefore a lower stabilisation energy. For borazine, there is now 3 nitrogens and 3 borons, this will lead to an even poorer overlap as these elements have a lager difference than each individually would have with carbon. For similar reasons as touched on, the stabilisation of each molecule will vary, with benzene having the greatest stabilisation energy, due to greater overlap of orbitals. This will give the largest energy difference between benzene and borazine due to the larger gap btween boron and nitrogen.
The degeneracy of the MOs, will be highest in benzene, as degeneracy is related to symmetry and this has the highest symmetry. Borazine will have a lower symmetry due to it having 2 different atoms, however as the molecule still has some symmetry as all atoms were changed, the degeneracy will not be as bad as that of boratabenzene and pyridinium. The order of expected degeneracy is likley to be benzene, borazine, boratabenzene, and pyridinium.
References
<references /1>

- ↑ http://www.huntresearchgroup.org.uk/teaching/year3_lab_start.html
- ↑ Acta vol. 36a Pg125-135
- ↑ http://en.wikipedia.org/wiki/Borazine