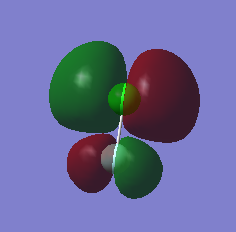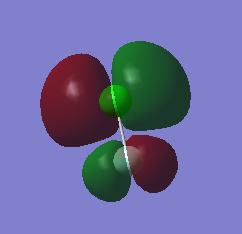Rep:Mod:Alexpaterson
Introduction
In this report the molecules NH3, N2, H2, ClF and Cl2 have been analysed using GaussView software. All these molecules were made and optimised and a summary information section on each molecule has been reported, along with their vibrational frequencies. For the molecules NH3 and ClF charge distribution was explored and explained using electronegativities. NH3 vibrational frequencies were observed in further detail. Molecular orbital analysis and explanation was also undertaken upon the ClF molecule.
Using the energies calculated from NH3, N2 and H2 a calculation was made in order to determine the reaction energies of the Haber-Bosch process.
NH3 Molecule
NH3 Molecule |
Summary information
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -56.55776873 au |
| RMS gradient | 0.00000485 |
| Point group | C3V |
| N-H bond length | 1.01798 au |
| H-N-H bond angle | 105.741 Degrees |
Item table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986274D-10
Optimization completed.
-- Stationary point found.
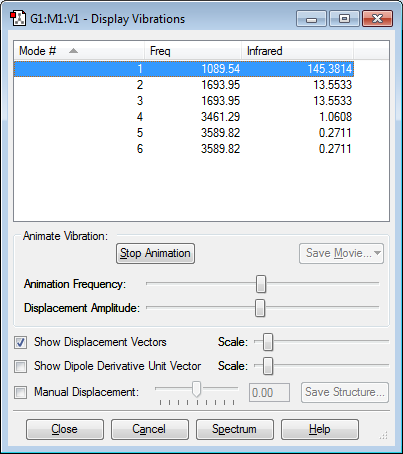
Vibrations display
This confirms that the molecule is at a minimum optimisation point as there are no negative frequencies observed.
Modes expected: 6
Degenerate modes: 2 and 3, 5 and 6
Bending vibration modes: 1, 2 and 3
Stretching vibration modes: 4, 5 and 6
Highly symmetric mode: 4
Umbrella mode: 1
How many modes expected to be seen in an experimental spectrum of gaseous ammonia: 2
Only two modes are high enough in intensity to be observed. These are 1 and 2/3 (which are degenerate in energy). Modes 4, 5 and 6 are too low in intensity to be observed because they cause only a small change in the dipole moment.
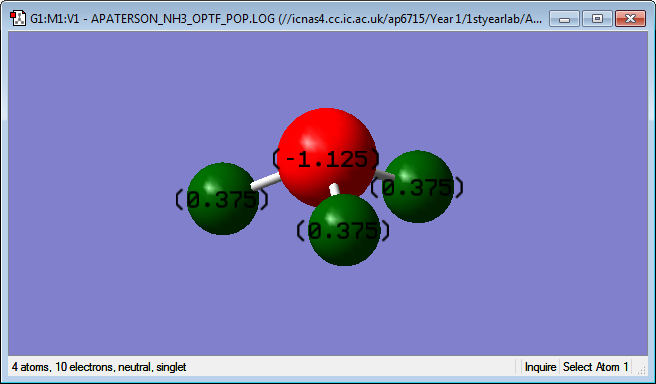
Charge distribution
The charge distribution, seen below, shows that the N atom has a value of -1.125 and the H atom has a value of 0.375. This is expected because N is a lot more electronegative than H and so it is expected that the bonding electrons are drawn towards it and, hence away from the H atoms. As electrons are negative the N atom will have a negative value and the H atom will have a positive value.
The log file can be found by this linkː Ammonia log file
N2 Molecule
N2 Molecule |
Summary information
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -109.52412868 au |
| RMS gradient | 0.00000060 |
| Point group | D infinity H |
| N-N bond length | 1.10550 au |
Item table
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401061D-13
Optimization completed.
-- Stationary point found.
Vibrations display
This confirms that the molecule is at a minimum optimisation point as there are no negative frequencies observed.
The log file can be found by this linkː Nitrogen log file
H2 Molecule
H2 Molecule |
Summary information
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -1.17853936 au |
| RMS gradient | 0.00000017 |
| Point group | D infinity H |
| H-H bond length | 0.74279 au |
Item table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
Vibrations display
This confirms that the molecule is at a minimum optimisation point as there are no negative frequencies observed.
The log file can be found by this linkː Hydrogen log file
Haber-Bosch process reaction energies
E(NH3)= -56.55776873 au 2*E(NH3)= -113.1155375 au E(N2)= -109.52412868 au E(H2)= -1.17853936 au 3*E(H2)= -3.53561808 au ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -146.48 KJ/mol
The ammonia product is more stable as the reaction is exothermic. This means that the ammonia product is at a lower energy and, hence is more stable than the gaseous products.
ClF Molecule
ClF Molecule |
Summary information
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -559.94269578 au |
| RMS gradient | 0.00014211 |
| Point group | C infinity V |
| Cl-F bond length | 1.66434 au |
Item table
Item Value Threshold Converged?
Maximum Force 0.000246 0.000450 YES
RMS Force 0.000246 0.000300 YES
Maximum Displacement 0.000433 0.001800 YES
RMS Displacement 0.000613 0.001200 YES
Predicted change in Energy=-1.066053D-07
Optimization completed.
-- Stationary point found.
Vibrations display
This confirms that the molecule is at a minimum optimisation point as there are no negative frequencies observed.
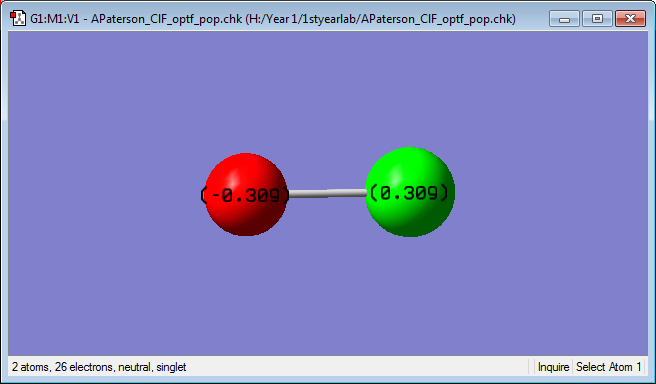
Charge distribution
The charge distribution, seen below, shows that the F atom has a value of -0.309 and the Cl atom has a value of 0.309. This is expected because F is more electronegative than Cl and so it is expected that the bonding electrons are drawn towards it and, hence away from the Cl atom. As electrons are negative the F atom will have a negative value and the Cl atom will have a positive value.
Molecular orbitals
MO 14, seen below, is the lowest unoccupied molecular orbital (LUMO) of ClF. It is the only MO that I have looked at that is not occupied by electrons. It is formed by the end on end out of phase overlap of the 2pz orbital of the F atom and the 3pz orbital of the Cl atom. They have a sigma star symmetry. This molecular orbital is an anti-bonding orbital because it is an out of phase overlap. It can be seen that there is less electron density between the two atoms compared to the in phase overlap seen by MO9 (The bonding-anti-bonding pair orbital). The contribution to the MO is larger by the Cl atom because it is the least electronegative of the two elements. This means that the 3pz orbital energy of Cl will be higher, and therefore closer to the molecular orbital than the 2pz of F, hence it will contribute more. The energy of this MO is -0.12150 au. This is the least deep in energy compared to all the other MO's that have been examined.
MO 13 and 12, both seen below, are degenerate. This is because they occupy the same energy level at -0.32855 au. These two MO's are known as the highest occupied molecular orbitals (HOMO's) of ClF. They are occupied with electrons and are the least deep in energy of all the occupied molecular orbitals of ClF. These MO's are formed via the side by side overlap of the 2py orbital of F and 3py orbital of Cl and the other degenerate orbital is formed by the side ways overlap of the 2px orbital of F and the 3px orbital of Cl. The MO's that are the bonding pair to these orbitals are MO 10 and 11. The symmetry of both these orbitals is pi star. These molecular orbitals are anti-bonding orbitals because they involve an out of phase overlap. There is a larger contribution to the MO's from the Cl atom because it is the least electronegative of the two elements. This means that the 3py and 3px orbital energy of Cl will be higher, and therefore closer to the molecular orbital than the 2py and 2px of F, and hence will contribute more.
MO9, seen below, is an occupied valence orbital of ClF. The energy of this MO is -0.52314 au. It is formed by the end on end in phase overlap of the 2pz orbital of F and the 3pz orbital of Cl. They have a sigma symmetry. This molecular orbital is a bonding orbital because it is an in phase overlap. There is more electron density between the two atoms when compared to MO 14 (the anti-bonding pair orbital), this shows it is a bonding orbital. There is also larger contribution from the F atom because the molecular orbital is a bonding orbital and F is the more electronegative element of the two atoms. This means that the 2pz orbital energy of F will be lower, and therefore closer to the molecular orbital than the 3pz of Cl, and hence will contribute more.
MO8, seen below, is also an occupied valence orbital of ClF. The energy of this MO is -0.83311. It is formed by the end on end out of phase overlap of the 2s orbital of F and the 3s orbital of Cl. They have a sigma star symmetry. This molecular orbital is an anti-bonding orbital because it is an out of phase overlap. There is less electron density between the two atoms when compared to MO 7 (the bonding pair orbital), this is because it is an anti bonding orbital. There is a larger contribution from the Cl atom because Cl is the least electronegative of the two atoms. This means that the 3s orbital energy of Cl will be higher, and therefore closer to the molecular orbital than the 2s of F, and hence will contribute more.
MO7, seen below, is also an occupied valence orbital of ClF. The energy of this MO is -1.21864 au. It is formed by the end on end in phase overlap of the 2s orbital of F and the 3s orbital of Cl. They have a sigma symmetry. This molecular orbital is a bonding orbital because it is an in phase overlap. There is more electron density between the two atoms when compared to MO 8 (the anti-bonding pair orbital), this is because it is a bonding orbital. There is a larger contribution from the F atom because F is the most electronegative element of the two atoms. This means that the 2s orbital energy of F will be lower, and therefore closer to the molecular orbital than the 3s of Cl, and hence will contribute more. Out of all the MO's examined this is the deepest in energy.
When choosing the MO's only valence MO's were observed because the other MO's are too deep in energy to get involved with the bonding. There are two types of orbitals, anti-bonding and bonding. If electrons occupy an anti-bonding orbital then they are destructive, if they are in a bonding orbital they are constructive. The overall bonding order can be calculated from the electron occupancy of the valence orbitals. There are 14 valence electrons. 7 from the F atom (2s2 2p5) and 7 from the Cl atom (3s2 3p5). There are 8 electrons contained within valence bonding MO's (MO7, MO9, MO10 and MO11) and there are 6 electrons contained within anti-bonding valence MO's (MO8, MO12 and MO13). Therefore the overall bond order is (8-6)/2=1. This means that there is only a single bond observed between the Cl and F atom.
The log file can be found by this linkː Chlorine monoflouride log file
Cl2 Molecule
Cl2 Molecule |
Summary information
| Calculation method | RB3LYP |
| Basis set | 6-31G(d,p) |
| Final energy E(RB3LYP) | -920.34987886 au |
| RMS gradient | 0.00002511 |
| Point group | D infinity H |
| Cl-Cl bond length | 2.04174 au |
Item table
Item Value Threshold Converged?
Maximum Force 0.000043 0.000450 YES
RMS Force 0.000043 0.000300 YES
Maximum Displacement 0.000121 0.001800 YES
RMS Displacement 0.000172 0.001200 YES
Predicted change in Energy=-5.277303D-09
Optimization completed.
-- Stationary point found.
Vibrations display
This confirms that the molecule is at a minimum optimisation point as there are no negative frequencies observed.
The log file can be found by this linkː Chlorine log file