Rep:Mod:AN910105
Introduction
Why do computational chemistry
For discovering the structure and bonding of molecules, it is relatively straight forward in organic system. However, it is not the case for inorganic system. In order to overcome this problem, the computational chemistry was introduced around 1930. By using the correlated chemistry theory as well as suitable computer programs, it gives us a significant amount of information about complexes' structure.The position of the transition states(TS) or active species can be determined, which can be difficult to observe experimentally.The energy of stable states are used to analysis thermodynamic data while the kinetic data can be obtained from the energy of TS.
What programme is used
GaussView 5.0.9 is used here.
Optimisation
BH3 Optimisation
BH3 Optimisation(Calculation Method: RB3LYP,Basis Set:3-21G)
Before optimistation:
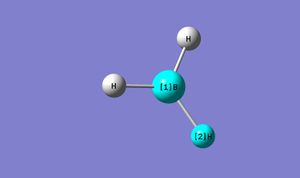
The bond length of B-H bond is manually altered to 1.50Å
After optimisation:
The log file of optimisation:click here
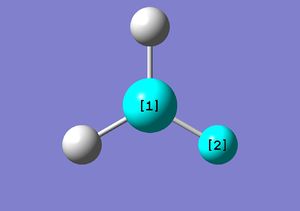
| BH3 Optimisation(Basis Set:3-21G) | ||
| File Name | BH3 | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 3-21G | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.46226338 | a.u. |
| RMS Gradient Norm | 0.00020672 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 27.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
From the data obtained above, it is clearly stated that the structure of the molecule is D3H, trigonal planar.The optimised B-H bond length is 1.193Å. The angel between two B-H bond is 120°
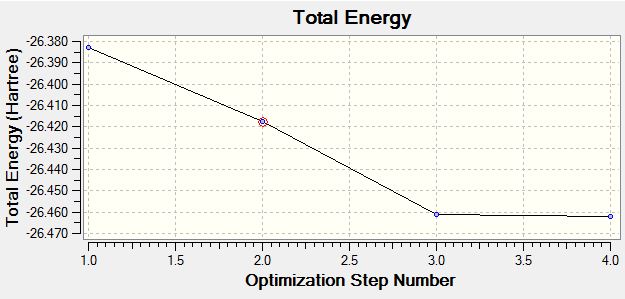
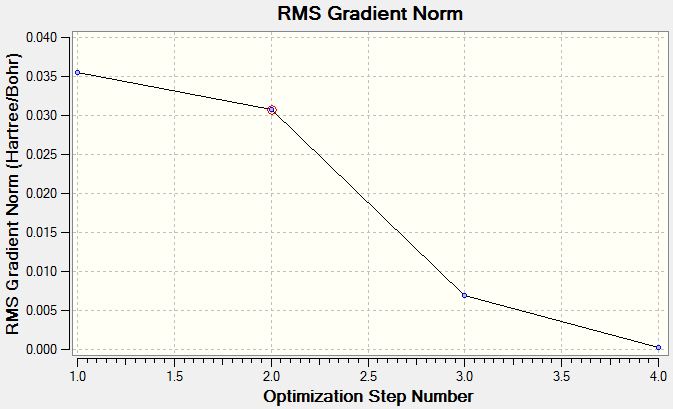
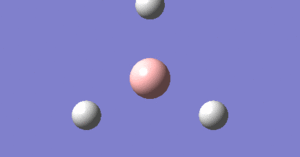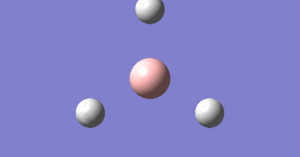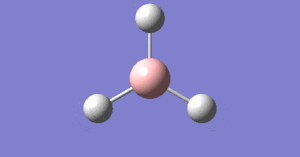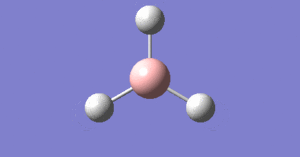
The energy change and gradient change over the optimisation
The following graph describes how the potential energy changes during the optimisation. the horizontal axis is the optimization step number and the vertial axis is the potential energy in hartree.
The graph below tells us how the energy gradient changes for each optimisation step. Th gradient unit is hartree/Bohr.
The optimisation is the process of finding a potential energy minimum which is also referred as a equilibrium position of nuclear and electron by changing the molecule slightly, in this case the bond length has been adjusted.According to the graph, a potential minimum can be found when gradient is zero, in other words, the energy of the molecule does not change respect to different optimisations.
BH3 Optimisation(Calculation Method: RB3LYP,Basis Set:6-31G(d,p))
Before optimistation:
The bond length of B-H bond is 1.193Å
After optimisation:
The log file of optimisation: click here
| BH3 Optimisation(Basis Set:6-31G(d,p)) | ||
| File Name | BH3_6-31G_before | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.61532363 | a.u. |
| RMS Gradient Norm | 0.00000235 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 32.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1923 -DE/DX = 0.0 !
! R2 R(1,3) 1.1923 -DE/DX = 0.0 !
! R3 R(1,4) 1.1923 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
From the data obtained above, it is also stated that the structure of the molecule is D3H, trigonal planar.The optimised B-H bond length is 1.192Å. The optimisd H-B-H bond angle is 120°,which is exactly the same as the data obtained from the optimisation using basis set 3-21G.
Comparison of two BH3 Optimisations
In terms of total energy of two different optimised molecule, the comparison has been made,which is shown below.
| final energy difference of BH3 Optimisation | ||
| basis set | energy/a.u. | |
| 3-21G | -26.46226338 | |
| 6-31G(d,p) | -26.61532363 | |
| energy difference:0.15306025 a.u. | ||
As you can see, the energy difference is around 402kJ/mol, which is fairly significant amount. The optimisation using basis set 6-31G(d,p)has a lower energy.
Optimisation of TIBr3
The D space calculation is provided:[| click here]
The log file of optimisation:click here

| TIBr3 Optimisation | ||
| File Name | TIBr3_optimisation | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | LANL2DZ | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -91.21812851 | a.u. |
| RMS Gradient Norm | 0.00000090 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 0 minutes 59.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084008D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.651 -DE/DX = 0.0 !
! R2 R(1,3) 2.651 -DE/DX = 0.0 !
! R3 R(1,4) 2.651 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
From the data obtained above, the structure of the molecule is D3H, trigonal planar.The optimised TI-Br bond length is 2.65Å. The optimisd Br-TI-Br bond angle is 120°
The literature optimised Tl-Br bond distance is 2.71Å, It is more or less the same as calculated value
Optimisation of BBr3
The log file of optimisation:click here

| BBr3 Optimisation | ||
| File Name | BBr3_6-31G | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | Gen | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -64.43645296 | a.u. |
| RMS Gradient Norm | 0.00000382 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours 1 minutes 36.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.026911D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.934 -DE/DX = 0.0 !
! R2 R(1,3) 1.934 -DE/DX = 0.0 !
! R3 R(1,4) 1.934 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
according to the item data, the optimisation is converged. Moreover,the structure of the molecule is D3H, trigonal planar.The optimised B-Br bond length is 1.93Å, three bonds are identical to each other.The optimisd Br-B-Br bond angle is 120°,three angles are identical.
Comparison of Bond Lengths and Bond Angles
After the optimisation of three different molecule which are all having the same symmetry,D3H,the bond length and bong angle have been analysed.
| Bond Lengths and Bond Angles for BH3,TIBr3,BBr3 | ||
| Molecule | Bond Length /Å | Bond Angle/degree |
|---|---|---|
| BH3 | 1.193 | 120.0 |
| TIBr3 | 2.65 | 120.0 |
| BBr3 | 1.93 | 120.0 |
As we can see from the table the bond angles are the same for three molecules. This tells us the 3-D structure of the molecule remains the same even the bonding atoms are changing.
However ,the bond length is changing along with the change in bonding orbital. Compared to BBr3 , BH3 has a shorter bond length due to the missing lone pair of electrons on the hydrogen which can donate to the p orbital of central boron Br has a lone pair of electrons that can donate, so better bonding between B and Br. For TI and B, the bonding orbitals of TI is larger in size, which results in poorer overlap between center TI atom and hydrogen.So TI-bond is longer than B-Br bond.
As gaussview shows the bond with in a range, sometimes the bond can not be shown as we expected but it does not mean there is no bonding between the atoms. The bond is a attractive interaction of 2 adjacent atoms. Covalent bond is two bonding share electron while the ionic bonding is the electrostatic attractions between two opposite charges.
Frequency Analysis and Association Energy
Frequency analysis for BH3
The log file of the frequency analysis:click here

| BH3 frequency analysis | ||
| File Name | FOR_FREQUNCY_BH3_6_31G_NEW | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -26.61532363 | a.u. |
| RMS Gradient Norm | 0.00000480 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 0.0000 | Debye |
| Point Group | C3H | |
| Job cpu time: 0 days 0 hours o minutes 35.0 seconds. | ||
The item table from the "real" output is showing below.
Item Value Threshold Converged?
Maximum Force 0.000010 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000038 0.001800 YES
RMS Displacement 0.000019 0.001200 YES
Predicted change in Energy=-5.370783D-10
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The low frequency is recorded in the text form file:
Low frequencies --- -3.6018 -1.1356 -0.0054 1.3735 9.7036 9.7698 Low frequencies --- 1162.9825 1213.1733 1213.1760
It is fairly clear that each molecule has a 3N-6 vibration frequencies,the frequencies listed above are the "-6",which describes the motions of the center of mass of the molecule. Normally, these 6 frequencies are within a range of -15cm-1to+15cm-1. As we can see from the data above, the 6 frequencies of this calculation is within a range of -4cm-1 to 10cm-1. This gives us a indication that the frequency calculation has been carried out correctly.
The following table summary all 6 vibration motions with their relative frequencies and individual intensity.
The predicted IR spectrum is showing below:
It is obvious that there are no 6 different peak for those vibration motion mentioned above. In reality, there are only 3 major peaks,whose corresponding frequencies are around 1150,1200,2700cm-1 respectively.
The reason for this is:
a)some of the vibration motions's frequencies are too close each, almost the same if showing graphical.This results in that only a single IR peak represents two motion.Back to the table above, vibration motion 2(scissoring) and 3(rocking) show a peak at about 1213cm-1 while vibration motion 5(asymmetry stretching) and 6(asymmetry stretching) show a peak at about 2715cm-1
b)one of the motions is predicted to have zero intensity, in other word, there will be no absorbancce peak for that vibration motion. In this case, motion 4(symmetry stretching)dose not have a IR peak in the spectrum.
Frequency analysis for TIBr3
The D space calculation is provided:[| click here]
The log file of the frequency analysis:click here
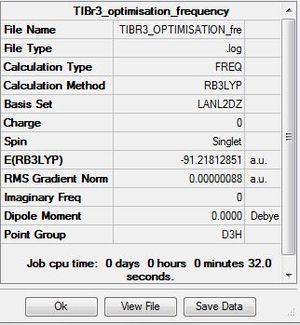
| TIBr3 frequency analysis | ||
| File Name | TIBR3_OPTIMISATION_fre | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | LANL2DZ | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -91.21812851 | a.u. |
| RMS Gradient Norm | 0.00000088 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 0.0000 | Debye |
| Point Group | D3H | |
| Job cpu time: 0 days 0 hours o minutes 32.0 seconds. | ||
The item table from the "real" output is showing below.
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The low frequency is recorded in the text form file:
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367 Low frequencies --- 46.4289 46.4292 52.1449
The predicted IR spectrum is showing below:
Comparison between Vibrational Frequencies of BH3 and TlBr3
After the two frequency calculation for these two molecules, analysis has been done to discover the nature of two molecule in terms of frequency,vibration modes and IR spectrum.
| Vibration motion number | Table (10) BH3 frequency/cm-1,intensity | TlBr BBr3 frequency/cm-1,intensity |
|---|---|---|
| 1 | 1162.98, 92.55 | 46.43, 3.6867 |
| 2 | 1213.17, 14.05 | 46.43, 3.6867 |
| 3 | 1213.18, 14.06 | 52.14, 5.8466 |
| 4 | 2582.32, 0.00 | 165.27, 0.0000 |
| 5 | 2715.50, 126.33 | 210.69, 25.4830 |
| 6 | 2715.50, 126.32 | 210.69, 25.4797 |
As we can see from the table above, all the frequencies for B-H bond is bigger than that of TI-Br bond. The difference in the reduced mass of two bond cause the reduce in frequency. TIBr3 is heavier than BH3 and absorbs more energy to vibrate. Recording of modes,A2’,E’ is recorded. Although the the frequency depends on the reduced mass of bonding atoms, the change in each vibration motion will be slightly different. For both molecules, they all have 6 vibration modes but all have only 3 major IR peak.
Using the same calculation method and basis set ensure all the calculations have been carried out at the same level of accuracy and the same possibility of finding error in the results. Threre is a energy different of 400kj/mol between the calculation basis 3-3G-21 and 6G-31(d,p). If method and basis sets of two calculation are different, the accuracy and possibility of find error will be different, which would leads the comparison to be incorrect.
The frequency analysis helps us to find the minimum of the reaction. As it is the derivative of the potential energy surface, negative frequencies represents a graphical maximum which is corresponding the transition state for the reaction.
The low frequencies are the indication of whether the optimisation is successful. If the value is negative, this indicates the calculation is failed to find the minimum and it usually requires better basis set to overcome the problem.
Molecular Orbitals of BH3
Not only the frequencies of vibration modes can be calculated, but also the graphical MO orbital is readily obtained if energy calculation which is also called population calculation is carried out under full NBO. It is essential to full in "pop=full" in to additional keywords section for MO analysis.
The D space calculation is provided:[| click here]
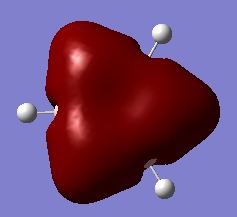
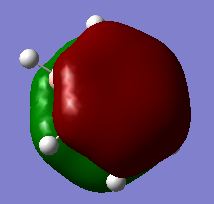
Calculated Molecular Orbital Diagram of BH3
There is no significant differentce between the real and LCAO MOs. The calculation is fairly accurate for small molecule like BH3.
Analysis of NH3
Optimisation of NH3
Before optimistation:
N-H bond is 1.00Å and the H-N-H angle is 109.47°
After optimisation:
The optimisation calculation is carried out under those conditions: job type is optimization, method used is B3LYP, basis set is 6-31G(d,p), additional keyword is nosymm.
The log file of optimisation:click here
| NH3 Optimisation | ||
| File Name | NH_3_ORIGINAL | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -56.54794758 | a.u. |
| RMS Gradient Norm | 0.00000975 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 1.9121 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 1 minutes 18.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000016 0.000450 YES
RMS Force 0.000013 0.000300 YES
Maximum Displacement 0.000122 0.001800 YES
RMS Displacement 0.000074 0.001200 YES
Predicted change in Energy=-2.899436D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.0194 -DE/DX = 0.0 !
! R2 R(1,3) 1.0194 -DE/DX = 0.0 !
! R3 R(1,4) 1.0194 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7493 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7607 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.76 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8821 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
According to the calculation above, the optimised bond length is 1.02Å,the predicted H-N-H angle is 105.76°,three angels are identical.
Frequency analysis of NH3
The frequency calculation is carried out under those conditions: job type is frequency, method used is B3LYP, basis set is 6-31G(d,p).
The log file of the frequency analysis:click here
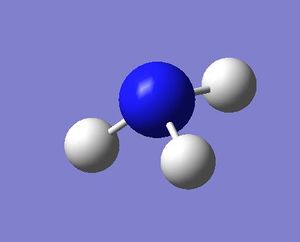
| NH3 frequency analysis | ||
| File Name | NH_3_ORIGINAL_FOR_FRE | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -56.55776856 | a.u. |
| RMS Gradient Norm | 0.00000480 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 0.00000888 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours o minutes 31.0 seconds. | ||
The item table from the "real" output is showing below.
Item Value Threshold Converged?
Maximum Force 0.000021 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000077 0.001800 YES
RMS Displacement 0.000039 0.001200 YES
Predicted change in Energy=-1.603126D-09
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
The low frequency is recorded in the text form file:
Low frequencies --- -30.7764 -0.0014 -0.0011 0.0013 20.3142 28.2484 Low frequencies --- 1089.5557 1694.1237 1694.1868
Population analysis of NH3
The population calculation is carried out under those conditions: job type is energy, method used is B3LYP, basis set is 6-31G(d,p).The form of the input file should be the checkpoint .chk file, not the log file.
The D space calculation is provided:[| click here]
The log file of the population analysis:click here

| NH3 population analysis | ||
| File Name | NH_3_ORIGINAL_POLULATION | |
| File Type | .log | |
| Calculation Type | SP | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -56.55776338 | a.u. |
| RMS Gradient Norm | a.u. | |
| Imaginary Freq | ||
| Dipole Moment | 1.8456 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours o minutes 13.0 seconds. | ||
The following figure lists the lowest 8 molecular orbital:

NMO analysis of NH3
By looking at charge distribution of the calculation results,the charge distribution can be visualized in terms of colour change as well as number

Quantitative data is recorded in the log file of the calculation. All sort of information about the molecule can be found out, for example, the bonding in the compound,interactions between the various orbitals including mixing orbitals as well as the energy and population or occupation of the N-H bonds and the nitrogen lone pair
Summary of Natural Population Analysis: Natural Population Natural ----------------------------------------------- Atom No Charge Core Valence Rydberg Total ----------------------------------------------------------------------- N 1 -1.12578 1.99982 6.11176 0.01421 8.12578 H 2 0.37526 0.00000 0.62228 0.00246 0.62474 H 3 0.37526 0.00000 0.62228 0.00246 0.62474 H 4 0.37527 0.00000 0.62228 0.00246 0.62473 ======================================================================= * Total * 0.00000 1.99982 7.97860 0.02158 10.00000
(Occupancy) Bond orbital/ Coefficients/ Hybrids --------------------------------------------------------------------------------- 1. (1.99909) BD ( 1) N 1 - H 2 ( 68.84%) 0.8297* N 1 s( 24.83%)p 3.02( 75.08%)d 0.00( 0.09%) -0.0001 -0.4983 -0.0059 0.0000 -0.2915 0.0052 0.8155 0.0275 0.0000 0.0000 0.0281 0.0000 0.0000 0.0032 0.0082 ( 31.16%) 0.5582* H 2 s( 99.91%)p 0.00( 0.09%) -0.9996 0.0000 0.0072 -0.0290 0.0000 2. (1.99909) BD ( 1) N 1 - H 3 ( 68.84%) 0.8297* N 1 s( 24.83%)p 3.02( 75.08%)d 0.00( 0.09%) 0.0001 0.4983 0.0059 0.0000 0.2915 -0.0052 0.4077 0.0138 0.7062 0.0239 0.0140 0.0243 0.0076 0.0034 0.0031 ( 31.16%) 0.5582* H 3 s( 99.91%)p 0.00( 0.09%) 0.9996 0.0000 -0.0072 -0.0145 -0.0251 3. (1.99909) BD ( 1) N 1 - H 4 ( 68.84%) 0.8297* N 1 s( 24.83%)p 3.02( 75.08%)d 0.00( 0.09%) 0.0001 0.4983 0.0059 0.0000 0.2915 -0.0053 0.4077 0.0138 -0.7062 -0.0239 0.0140 -0.0243 -0.0076 0.0034 0.0031 ( 31.16%) 0.5582* H 4 s( 99.91%)p 0.00( 0.09%) 0.9996 0.0000 -0.0072 -0.0145 0.0251 4. (1.99982) CR ( 1) N 1 s(100.00%) 1.0000 -0.0002 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 5. (1.99720) LP ( 1) N 1 s( 25.48%)p 2.92( 74.42%)d 0.00( 0.10%) 0.0001 0.5046 -0.0120 0.0000 -0.8612 0.0502 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 0.0000 -0.0268 0.0155 6. (0.00000) RY*( 1) N 1 s( 99.98%)p 0.00( 0.02%)d 0.00( 0.00%) 7. (0.00000) RY*( 2) N 1 s(100.00%) 8. (0.00000) RY*( 3) N 1 s( 0.03%)p99.99( 99.97%)d 0.01( 0.00%) 9. (0.00000) RY*( 4) N 1 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%) 10. (0.00000) RY*( 5) N 1 s( 0.00%)p 1.00(100.00%)d 0.00( 0.00%) 11. (0.00000) RY*( 6) N 1 s( 0.00%)p 1.00( 0.12%)d99.99( 99.88%) 12. (0.00000) RY*( 7) N 1 s( 0.00%)p 1.00( 0.12%)d99.99( 99.88%) 13. (0.00000) RY*( 8) N 1 s( 0.00%)p 1.00( 0.01%)d99.99( 99.99%) 14. (0.00000) RY*( 9) N 1 s( 0.01%)p 4.40( 0.06%)d99.99( 99.92%) 15. (0.00000) RY*(10) N 1 s( 0.00%)p 1.00( 0.03%)d99.99( 99.97%) 16. (0.00112) RY*( 1) H 2 s( 72.77%)p 0.37( 27.23%) 0.0039 0.8530 0.5218 -0.0021 -0.0002 17. (0.00045) RY*( 2) H 2 s( 26.60%)p 2.76( 73.40%) -0.0018 0.5158 -0.8437 -0.1490 -0.0011 18. (0.00034) RY*( 3) H 2 s( 0.00%)p 1.00(100.00%) 0.0000 0.0007 -0.0008 -0.0001 1.0000 19. (0.00000) RY*( 4) H 2 s( 0.72%)p99.99( 99.28%) 20. (0.00112) RY*( 1) H 3 s( 72.77%)p 0.37( 27.23%) 0.0039 0.8530 0.5218 0.0012 0.0017 21. (0.00045) RY*( 2) H 3 s( 26.60%)p 2.76( 73.40%) -0.0018 0.5158 -0.8437 0.0754 0.1285 22. (0.00034) RY*( 3) H 3 s( 0.00%)p 1.00(100.00%) 0.0000 0.0006 -0.0008 -0.8660 0.5001 23. (0.00000) RY*( 4) H 3 s( 0.72%)p99.99( 99.28%) 24. (0.00112) RY*( 1) H 4 s( 72.75%)p 0.37( 27.25%) 0.0039 0.8529 0.5220 0.0010 -0.0018 25. (0.00044) RY*( 2) H 4 s( 26.62%)p 2.76( 73.38%) -0.0017 0.5159 -0.8436 0.0744 -0.1290 26. (0.00034) RY*( 3) H 4 s( 0.00%)p 1.00(100.00%) 0.0000 0.0000 0.0000 0.8660 0.5000 27. (0.00000) RY*( 4) H 4 s( 0.72%)p99.99( 99.28%) 28. (0.00000) BD*( 1) N 1 - H 2 ( 31.16%) 0.5582* N 1 s( 24.83%)p 3.02( 75.08%)d 0.00( 0.09%) ( 68.84%) -0.8297* H 2 s( 99.91%)p 0.00( 0.09%) 29. (0.00000) BD*( 1) N 1 - H 3 ( 31.16%) 0.5582* N 1 s( 24.83%)p 3.02( 75.08%)d 0.00( 0.09%) ( 68.84%) -0.8297* H 3 s( 99.91%)p 0.00( 0.09%) 30. (0.00000) BD*( 1) N 1 - H 4 ( 31.16%) 0.5582* N 1 s( 24.83%)p 3.02( 75.08%)d 0.00( 0.09%) ( 68.84%) -0.8297* H 4 s( 99.91%)p 0.00( 0.09%)
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis Threshold for printing: 0.50 kcal/mol E(2) E(j)-E(i) F(i,j) Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u. ===================================================================================================
Natural Bond Orbitals (Summary): Principal Delocalizations NBO Occupancy Energy (geminal,vicinal,remote) ==================================================================================== Molecular unit 1 (H3N) 1. BD ( 1) N 1 - H 2 1.99909 -0.60344 2. BD ( 1) N 1 - H 3 1.99909 -0.60344 3. BD ( 1) N 1 - H 4 1.99909 -0.60344 4. CR ( 1) N 1 1.99982 -14.16853 5. LP ( 1) N 1 1.99720 -0.31788 24(v),16(v),20(v),17(v) 21(v),25(v) 6. RY*( 1) N 1 0.00000 1.20608 7. RY*( 2) N 1 0.00000 3.73049 8. RY*( 3) N 1 0.00000 0.73752 9. RY*( 4) N 1 0.00000 0.77372 10. RY*( 5) N 1 0.00000 0.77373 11. RY*( 6) N 1 0.00000 2.28867 12. RY*( 7) N 1 0.00000 2.28871 13. RY*( 8) N 1 0.00000 2.40772 14. RY*( 9) N 1 0.00000 2.16194 15. RY*( 10) N 1 0.00000 2.32578 16. RY*( 1) H 2 0.00112 1.11339 17. RY*( 2) H 2 0.00045 1.84782 18. RY*( 3) H 2 0.00034 2.31860 19. RY*( 4) H 2 0.00000 2.94410 20. RY*( 1) H 3 0.00112 1.11338 21. RY*( 2) H 3 0.00045 1.84783 22. RY*( 3) H 3 0.00034 2.31860 23. RY*( 4) H 3 0.00000 2.94409 24. RY*( 1) H 4 0.00112 1.11374 25. RY*( 2) H 4 0.00044 1.84748 26. RY*( 3) H 4 0.00034 2.31858 27. RY*( 4) H 4 0.00000 2.94404 28. BD*( 1) N 1 - H 2 0.00000 0.48363 29. BD*( 1) N 1 - H 3 0.00000 0.48361 30. BD*( 1) N 1 - H 4 0.00000 0.48359
Association energy analysis of product NH3BH3
Ammonia-borane has a high hydrogen content and is stable at standard condition. What is even more, it is acid-base complex. This makes the molecule to be very useful in terms of fuel.To determine the reaction energy,both reactant and product's energy is required.AS calculation has been done for both reactant,we can obtain the reaction energy quite simply.
As the molecule has more than 3 atoms, 2 optimisation calculations are recommended, in which the calculation use method used,B3LYP,and basis set,3-21G, while the second one use method used,B3LYP,and basis set,6-31G(d,p).For second calculation, a additional keywords,nosymm,is required.
First optimisation
The log file of optimisation:click here

| NH3BH3 Optimisation | ||
| File Name | nh3bh3 | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 3-21G | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -82.76661837 | a.u. |
| RMS Gradient Norm | 0.00003006 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 5.8431 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 1 minutes 55.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000094 0.000450 YES
RMS Force 0.000030 0.000300 YES
Maximum Displacement 0.000419 0.001800 YES
RMS Displacement 0.000178 0.001200 YES
Predicted change in Energy=-5.742843D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0277 -DE/DX = -0.0001 !
! R2 R(2,7) 1.0277 -DE/DX = -0.0001 !
! R3 R(3,7) 1.0277 -DE/DX = 0.0 !
! R4 R(4,8) 1.212 -DE/DX = 0.0 !
! R5 R(5,8) 1.212 -DE/DX = -0.0001 !
! R6 R(6,8) 1.212 -DE/DX = -0.0001 !
! R7 R(7,8) 1.6854 -DE/DX = -0.0001 !
! A1 A(1,7,2) 109.3469 -DE/DX = 0.0 !
! A2 A(1,7,3) 109.35 -DE/DX = 0.0 !
! A3 A(1,7,8) 109.5925 -DE/DX = 0.0 !
! A4 A(2,7,3) 109.3462 -DE/DX = 0.0 !
! A5 A(2,7,8) 109.5973 -DE/DX = 0.0 !
! A6 A(3,7,8) 109.5935 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.563 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.5594 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.9872 -DE/DX = 0.0 !
! A10 A(5,8,6) 113.5575 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.9846 -DE/DX = 0.0 !
! A12 A(6,8,7) 104.9848 -DE/DX = 0.0 !
! D1 D(1,7,8,4) 179.9938 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -60.0024 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 59.994 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -60.0065 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 59.9972 -DE/DX = 0.0 !
! D6 D(2,7,8,6) 179.9936 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 59.9928 -DE/DX = 0.0 !
! D8 D(3,7,8,5) 179.9966 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -60.007 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Second optimisation
The log file of optimisation:click here
| NH3BH3 Second optimisation | ||
| File Name | nh3bh3_2OPTI | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -83.22469035 | a.u. |
| RMS Gradient Norm | 0.00005813 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 5.5628 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 1 minutes 37.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000137 0.000450 YES
RMS Force 0.000038 0.000300 YES
Maximum Displacement 0.001020 0.001800 YES
RMS Displacement 0.000225 0.001200 YES
Predicted change in Energy=-1.139077D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,7) 1.0185 -DE/DX = 0.0 !
! R2 R(2,7) 1.0185 -DE/DX = 0.0 !
! R3 R(3,7) 1.0185 -DE/DX = 0.0 !
! R4 R(4,8) 1.2097 -DE/DX = 0.0 !
! R5 R(5,8) 1.2097 -DE/DX = 0.0 !
! R6 R(6,8) 1.2097 -DE/DX = 0.0 !
! R7 R(7,8) 1.6685 -DE/DX = -0.0001 !
! A1 A(1,7,2) 107.8567 -DE/DX = 0.0 !
! A2 A(1,7,3) 107.8615 -DE/DX = 0.0 !
! A3 A(1,7,8) 111.0394 -DE/DX = 0.0 !
! A4 A(2,7,3) 107.861 -DE/DX = 0.0 !
! A5 A(2,7,8) 111.0396 -DE/DX = 0.0 !
! A6 A(3,7,8) 111.0358 -DE/DX = 0.0 !
! A7 A(4,8,5) 113.9015 -DE/DX = 0.0 !
! A8 A(4,8,6) 113.8953 -DE/DX = 0.0 !
! A9 A(4,8,7) 104.567 -DE/DX = 0.0001 !
! A10 A(5,8,6) 113.9013 -DE/DX = 0.0 !
! A11 A(5,8,7) 104.5643 -DE/DX = 0.0001 !
! A12 A(6,8,7) 104.5652 -DE/DX = 0.0001 !
! D1 D(1,7,8,4) -179.9977 -DE/DX = 0.0 !
! D2 D(1,7,8,5) -59.9951 -DE/DX = 0.0 !
! D3 D(1,7,8,6) 60.0064 -DE/DX = 0.0 !
! D4 D(2,7,8,4) -59.9999 -DE/DX = 0.0 !
! D5 D(2,7,8,5) 60.0027 -DE/DX = 0.0 !
! D6 D(2,7,8,6) -179.9958 -DE/DX = 0.0 !
! D7 D(3,7,8,4) 60.001 -DE/DX = 0.0 !
! D8 D(3,7,8,5) -179.9965 -DE/DX = 0.0 !
! D9 D(3,7,8,6) -59.995 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency analysis
The frequency calculation is carried out under those conditions: job type is frequency, method used is B3LYP, basis set is 6-31G(d,p).
The log file of the frequency analysis:click here
| NH3BH3 frequency analysis | ||
| File Name | nh3bh3_fre | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -83.22469035 | a.u. |
| RMS Gradient Norm | 0.00005810 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 5.5628 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 1 minutes 28.0 seconds. | ||
The item table from the "real" output is showing below.
Item Value Threshold Converged?
Maximum Force 0.000265 0.000450 YES
RMS Force 0.000058 0.000300 YES
Maximum Displacement 0.001477 0.001800 YES
RMS Displacement 0.000379 0.001200 YES
Predicted change in Energy=-2.171276D-07
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
low frequencies are showing below:
Low frequencies --- 0.0009 0.0010 0.0015 9.3420 10.9493 30.1266 Low frequencies --- 264.8040 631.3745 637.2810
Association Energy calculation
E(NH3)=-56.55776856a.u.
E(BH3)=-26.61532363a.u.
E(NH3BH3)=-83.22469035
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]=-83.22469035-[-56.55776856-26.61532363]=-0.05159816a.u. =-135.47kJ/mol
Mini Project-Investigating aromatically
Benzene
Optimisation
Job type: Optimisation Method: B3LYP Basis set: 6-31G(d,p) Additional keywords: nosymm
The log file of optimisation:click here
The D space calculation is provided:[| click here]

| Benzene optimisation | ||
| File Name | benzene | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -232.25821186 | a.u. |
| RMS Gradient Norm | 0.00009338 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0001 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 1 minutes 33.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000204 0.000450 YES
RMS Force 0.000084 0.000300 YES
Maximum Displacement 0.000870 0.001800 YES
RMS Displacement 0.000313 0.001200 YES
Predicted change in Energy=-4.983462D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3963 -DE/DX = 0.0001 !
! R2 R(1,6) 1.3961 -DE/DX = 0.0002 !
! R3 R(1,7) 1.0861 -DE/DX = 0.0002 !
! R4 R(2,3) 1.3961 -DE/DX = 0.0002 !
! R5 R(2,8) 1.0861 -DE/DX = 0.0002 !
! R6 R(3,4) 1.3963 -DE/DX = 0.0001 !
! R7 R(3,9) 1.0861 -DE/DX = 0.0002 !
! R8 R(4,5) 1.3961 -DE/DX = 0.0002 !
! R9 R(4,10) 1.0861 -DE/DX = 0.0002 !
! R10 R(5,6) 1.3963 -DE/DX = 0.0001 !
! R11 R(5,11) 1.0861 -DE/DX = 0.0002 !
! R12 R(6,12) 1.0861 -DE/DX = 0.0002 !
! A1 A(2,1,6) 119.9996 -DE/DX = 0.0 !
! A2 A(2,1,7) 119.9968 -DE/DX = 0.0 !
! A3 A(6,1,7) 120.0036 -DE/DX = 0.0 !
! A4 A(1,2,3) 120.0036 -DE/DX = 0.0 !
! A5 A(1,2,8) 119.9916 -DE/DX = 0.0 !
! A6 A(3,2,8) 120.0048 -DE/DX = 0.0 !
! A7 A(2,3,4) 119.9967 -DE/DX = 0.0 !
! A8 A(2,3,9) 120.0101 -DE/DX = 0.0 !
! A9 A(4,3,9) 119.9932 -DE/DX = 0.0 !
! A10 A(3,4,5) 119.9996 -DE/DX = 0.0 !
! A11 A(3,4,10) 119.9891 -DE/DX = 0.0 !
! A12 A(5,4,10) 120.0113 -DE/DX = 0.0 !
! A13 A(4,5,6) 120.004 -DE/DX = 0.0 !
! A14 A(4,5,11) 120.0048 -DE/DX = 0.0 !
! A15 A(6,5,11) 119.9912 -DE/DX = 0.0 !
! A16 A(1,6,5) 119.9965 -DE/DX = 0.0 !
! A17 A(1,6,12) 120.0072 -DE/DX = 0.0 !
! A18 A(5,6,12) 119.9963 -DE/DX = 0.0 !
! D1 D(6,1,2,3) -0.0059 -DE/DX = 0.0 !
! D2 D(6,1,2,8) 180.0021 -DE/DX = 0.0 !
! D3 D(7,1,2,3) -180.0099 -DE/DX = 0.0 !
! D4 D(7,1,2,8) -0.0019 -DE/DX = 0.0 !
! D5 D(2,1,6,5) -0.0055 -DE/DX = 0.0 !
! D6 D(2,1,6,12) -179.9972 -DE/DX = 0.0 !
! D7 D(7,1,6,5) -180.0016 -DE/DX = 0.0 !
! D8 D(7,1,6,12) 0.0068 -DE/DX = 0.0 !
! D9 D(1,2,3,4) 0.0119 -DE/DX = 0.0 !
! D10 D(1,2,3,9) 180.0087 -DE/DX = 0.0 !
! D11 D(8,2,3,4) 180.0039 -DE/DX = 0.0 !
! D12 D(8,2,3,9) 0.0007 -DE/DX = 0.0 !
! D13 D(2,3,4,5) -0.0064 -DE/DX = 0.0 !
! D14 D(2,3,4,10) -180.0058 -DE/DX = 0.0 !
! D15 D(9,3,4,5) 179.9968 -DE/DX = 0.0 !
! D16 D(9,3,4,10) -0.0026 -DE/DX = 0.0 !
! D17 D(3,4,5,6) -0.005 -DE/DX = 0.0 !
! D18 D(3,4,5,11) 180.0061 -DE/DX = 0.0 !
! D19 D(10,4,5,6) -180.0057 -DE/DX = 0.0 !
! D20 D(10,4,5,11) 0.0055 -DE/DX = 0.0 !
! D21 D(4,5,6,1) 0.011 -DE/DX = 0.0 !
! D22 D(4,5,6,12) 180.0027 -DE/DX = 0.0 !
! D23 D(11,5,6,1) 179.9999 -DE/DX = 0.0 !
! D24 D(11,5,6,12) -0.0085 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency Analysis
Job type: Frequency Method: B3LYP Basis set: 6-31G(d,p)
The log file of optimisation:click here
The D space calculation is provided:[| click here]
| Benzene frequency analysis | ||
| File Name | benzene_opt_fre_2 | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -232.25821186 | a.u. |
| RMS Gradient Norm | 0.00009343 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 0.0001 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 5 minutes 9.0 seconds. | ||
The item table from the "real" output is showing below.
Item Value Threshold Converged?
Maximum Force 0.000203 0.000450 YES
RMS Force 0.000093 0.000300 YES
Maximum Displacement 0.000834 0.001800 YES
RMS Displacement 0.000367 0.001200 YES
Predicted change in Energy=-4.855235D-07
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
low frequencies are showing below:
Low frequencies --- -14.2246 -2.7148 0.0006 0.0007 0.0010 10.0085 Low frequencies --- 413.7274 414.5533 621.0441
Population
Job type: Energy Method: B3LYP Basis set: 6-31G(d,p) Additional keywords: pop=full, NBO: Full NBO
The log file of the population analysis:click here
The D space calculation is provided:[| click here]
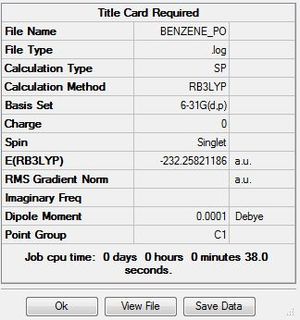
| Benzene population analysis | ||
| File Name | BENZENE_PO | |
| File Type | .log | |
| Calculation Type | SP | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -232.25821186 | a.u. |
| RMS Gradient Norm | a.u. | |
| Imaginary Freq | ||
| Dipole Moment | 0.0001 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours minutes 38.0 seconds. | ||
Pyridinium
Optimisation
Job type: Optimisation Method: B3LYP Basis set: 6-31G(d,p) Charge: +1
The log file of optimisation:click here
The D space calculation is provided:[| click here]
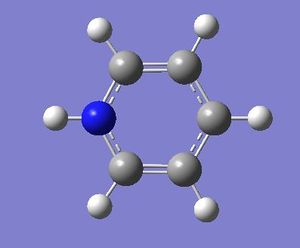
| Pyridinium optimisation | ||
| File Name | pyridinium | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| Spin | Singlet | |
| E(RB3LYP) | -248.66807396 | a.u. |
| RMS Gradient Norm | 0.00003896 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 1.8727 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 3 minutes 42.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged? Maximum Force 0.000064 0.000450 YES RMS Force 0.000023 0.000300 YES Maximum Displacement 0.000702 0.001800 YES RMS Displacement 0.000174 0.001200 YES Predicted change in Energy=-6.897705D-08 Optimization completed. -- Stationary point found. ---------------------------- ! Optimized Parameters ! ! (Angstroms and Degrees) ! -------------------------- -------------------------- ! Name Definition Value Derivative Info. ! -------------------------------------------------------------------------------- ! R1 R(1,2) 1.3839 -DE/DX = 0.0 ! ! R2 R(1,6) 1.0832 -DE/DX = 0.0 ! ! R3 R(1,12) 1.3523 -DE/DX = 0.0001 ! ! R4 R(2,3) 1.3988 -DE/DX = 0.0 ! ! R5 R(2,7) 1.0835 -DE/DX = 0.0 ! ! R6 R(3,4) 1.3988 -DE/DX = 0.0 ! ! R7 R(3,8) 1.0852 -DE/DX = 0.0 ! ! R8 R(4,5) 1.3838 -DE/DX = 0.0 ! ! R9 R(4,9) 1.0835 -DE/DX = 0.0 ! ! R10 R(5,10) 1.0832 -DE/DX = 0.0 ! ! R11 R(5,12) 1.3524 -DE/DX = 0.0 ! ! R12 R(11,12) 1.0169 -DE/DX = 0.0 ! ! A1 A(2,1,6) 123.9296 -DE/DX = 0.0 ! ! A2 A(2,1,12) 119.2363 -DE/DX = 0.0 ! ! A3 A(6,1,12) 116.8341 -DE/DX = 0.0 ! ! A4 A(1,2,3) 119.082 -DE/DX = 0.0 ! ! A5 A(1,2,7) 119.4193 -DE/DX = 0.0001 ! ! A6 A(3,2,7) 121.4987 -DE/DX = -0.0001 ! ! A7 A(2,3,4) 120.0549 -DE/DX = 0.0 ! ! A8 A(2,3,8) 119.974 -DE/DX = 0.0 ! ! A9 A(4,3,8) 119.9711 -DE/DX = 0.0 ! ! A10 A(3,4,5) 119.0827 -DE/DX = 0.0 ! ! A11 A(3,4,9) 121.4958 -DE/DX = -0.0001 ! ! A12 A(5,4,9) 119.4214 -DE/DX = 0.0 ! ! A13 A(4,5,10) 123.9324 -DE/DX = 0.0 ! ! A14 A(4,5,12) 119.2354 -DE/DX = 0.0 ! ! A15 A(10,5,12) 116.8322 -DE/DX = 0.0 ! ! A16 A(1,12,5) 123.3087 -DE/DX = 0.0 ! ! A17 A(1,12,11) 118.3463 -DE/DX = 0.0 ! ! A18 A(5,12,11) 118.345 -DE/DX = 0.0 ! ! D1 D(6,1,2,3) 180.0005 -DE/DX = 0.0 ! ! D2 D(6,1,2,7) 0.0001 -DE/DX = 0.0 ! ! D3 D(12,1,2,3) 0.0 -DE/DX = 0.0 ! ! D4 D(12,1,2,7) -180.0004 -DE/DX = 0.0 ! ! D5 D(2,1,12,5) 0.0018 -DE/DX = 0.0 ! ! D6 D(2,1,12,11) 180.0004 -DE/DX = 0.0 ! ! D7 D(6,1,12,5) 180.0013 -DE/DX = 0.0 ! ! D8 D(6,1,12,11) -0.0001 -DE/DX = 0.0 ! ! D9 D(1,2,3,4) -0.0007 -DE/DX = 0.0 ! ! D10 D(1,2,3,8) -179.9999 -DE/DX = 0.0 ! ! D11 D(7,2,3,4) -180.0003 -DE/DX = 0.0 ! ! D12 D(7,2,3,8) 0.0005 -DE/DX = 0.0 ! ! D13 D(2,3,4,5) -0.0002 -DE/DX = 0.0 ! ! D14 D(2,3,4,9) -179.9998 -DE/DX = 0.0 ! ! D15 D(8,3,4,5) -180.0011 -DE/DX = 0.0 ! ! D16 D(8,3,4,9) -0.0007 -DE/DX = 0.0 ! ! D17 D(3,4,5,10) -180.0007 -DE/DX = 0.0 ! ! D18 D(3,4,5,12) 0.0019 -DE/DX = 0.0 ! ! D19 D(9,4,5,10) -0.0011 -DE/DX = 0.0 ! ! D20 D(9,4,5,12) 180.0015 -DE/DX = 0.0 ! ! D21 D(4,5,12,1) -0.0028 -DE/DX = 0.0 ! ! D22 D(4,5,12,11) -180.0013 -DE/DX = 0.0 ! ! D23 D(10,5,12,1) -180.0003 -DE/DX = 0.0 ! ! D24 D(10,5,12,11) 0.0011 -DE/DX = 0.0 ! -------------------------------------------------------------------------------- GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency Analysis
Job type: Frequency Method: B3LYP Basis set: 6-31G(d,p) Charge:+1
The log file of the frequency analysis:click here
The D space calculation is provided:[| click here]

| Pyridinium frequency analysis | ||
| File Name | PYRIDINIUM_opt_fre | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| Spin | Singlet | |
| E(RB3LYP) | -248.66807396 | a.u. |
| RMS Gradient Norm | 0.00003889 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 1.8727 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 5 minutes 5.0 seconds. | ||
The item table from the "real" output is showing below.
Item Value Threshold Converged?
Maximum Force 0.000155 0.000450 YES
RMS Force 0.000039 0.000300 YES
Maximum Displacement 0.000774 0.001800 YES
RMS Displacement 0.000227 0.001200 YES
Predicted change in Energy=-7.429612D-08
Optimization completed.
-- Stationary point found.
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
low frequencies are showing below:
Low frequencies --- -7.2104 -0.0005 -0.0005 -0.0003 17.3406 18.5424 Low frequencies --- 392.4561 404.0617 620.4717
Population
Job type: Energy Method: B3LYP Basis set: 6-31G(d,p) Additional keywords: pop=full, NBO: Full NBO Charge: +1
The log file of the population analysis:click here
The D space calculation is provided:[| click here]
| Pyridinium population analysis | ||
| File Name | PYRIDINIUM_OPT_PO | |
| File Type | .log | |
| Calculation Type | SP | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 1 | |
| Spin | Singlet | |
| E(RB3LYP) | -248.66807396 | a.u. |
| RMS Gradient Norm | a.u. | |
| Imaginary Freq | ||
| Dipole Moment | 1.8727 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours minutes 43.0 seconds. | ||
Boratabenzene
Optimisation
Job type: Optimisation Method: B3LYP Basis set: 6-31G(d,p) Charge: -1
The log file of optimisation:click here
The D space calculation is provided:[| click here]
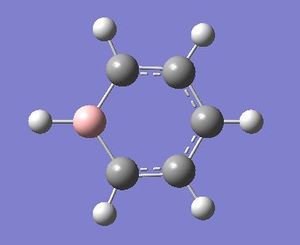
| Boratabenzene optimisation | ||
| File Name | Boratabenzene | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | -1 | |
| Spin | Singlet | |
| E(RB3LYP) | -219.02052984 | a.u. |
| RMS Gradient Norm | 0.00015822 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 11.0032 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 3 minutes 30.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000159 0.000450 YES
RMS Force 0.000069 0.000300 YES
Maximum Displacement 0.000911 0.001800 YES
RMS Displacement 0.000335 0.001200 YES
Predicted change in Energy=-6.630273D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3989 -DE/DX = 0.0 !
! R2 R(1,6) 1.097 -DE/DX = -0.0001 !
! R3 R(1,12) 1.5137 -DE/DX = 0.0001 !
! R4 R(2,3) 1.4053 -DE/DX = -0.0001 !
! R5 R(2,7) 1.0968 -DE/DX = 0.0001 !
! R6 R(3,4) 1.4053 -DE/DX = -0.0001 !
! R7 R(3,8) 1.0917 -DE/DX = -0.0001 !
! R8 R(4,5) 1.3989 -DE/DX = 0.0 !
! R9 R(4,9) 1.0968 -DE/DX = 0.0001 !
! R10 R(5,10) 1.097 -DE/DX = -0.0001 !
! R11 R(5,12) 1.5138 -DE/DX = 0.0001 !
! R12 R(11,12) 1.2185 -DE/DX = 0.0 !
! A1 A(2,1,6) 115.9495 -DE/DX = 0.0001 !
! A2 A(2,1,12) 120.0809 -DE/DX = -0.0001 !
! A3 A(6,1,12) 123.9696 -DE/DX = -0.0001 !
! A4 A(1,2,3) 122.1393 -DE/DX = 0.0001 !
! A5 A(1,2,7) 120.4236 -DE/DX = -0.0002 !
! A6 A(3,2,7) 117.437 -DE/DX = 0.0 !
! A7 A(2,3,4) 120.4508 -DE/DX = -0.0001 !
! A8 A(2,3,8) 119.7759 -DE/DX = 0.0001 !
! A9 A(4,3,8) 119.7732 -DE/DX = 0.0001 !
! A10 A(3,4,5) 122.1382 -DE/DX = 0.0001 !
! A11 A(3,4,9) 117.4352 -DE/DX = 0.0 !
! A12 A(5,4,9) 120.4267 -DE/DX = -0.0002 !
! A13 A(4,5,10) 115.9535 -DE/DX = 0.0001 !
! A14 A(4,5,12) 120.0812 -DE/DX = -0.0001 !
! A15 A(10,5,12) 123.9653 -DE/DX = -0.0001 !
! A16 A(1,12,5) 115.1096 -DE/DX = 0.0 !
! A17 A(1,12,11) 122.4483 -DE/DX = 0.0 !
! A18 A(5,12,11) 122.4422 -DE/DX = 0.0 !
! D1 D(6,1,2,3) 180.0103 -DE/DX = 0.0 !
! D2 D(6,1,2,7) -0.0041 -DE/DX = 0.0 !
! D3 D(12,1,2,3) 0.0085 -DE/DX = 0.0 !
! D4 D(12,1,2,7) -180.0059 -DE/DX = 0.0 !
! D5 D(2,1,12,5) 0.001 -DE/DX = 0.0 !
! D6 D(2,1,12,11) -179.9999 -DE/DX = 0.0 !
! D7 D(6,1,12,5) -180.0009 -DE/DX = 0.0 !
! D8 D(6,1,12,11) -0.0019 -DE/DX = 0.0 !
! D9 D(1,2,3,4) -0.015 -DE/DX = 0.0 !
! D10 D(1,2,3,8) -180.0107 -DE/DX = 0.0 !
! D11 D(7,2,3,4) -180.001 -DE/DX = 0.0 !
! D12 D(7,2,3,8) 0.0032 -DE/DX = 0.0 !
! D13 D(2,3,4,5) 0.0115 -DE/DX = 0.0 !
! D14 D(2,3,4,9) 180.0047 -DE/DX = 0.0 !
! D15 D(8,3,4,5) 180.0073 -DE/DX = 0.0 !
! D16 D(8,3,4,9) 0.0005 -DE/DX = 0.0 !
! D17 D(3,4,5,10) -180.0052 -DE/DX = 0.0 !
! D18 D(3,4,5,12) -0.0019 -DE/DX = 0.0 !
! D19 D(9,4,5,10) 0.0019 -DE/DX = 0.0 !
! D20 D(9,4,5,12) 180.0052 -DE/DX = 0.0 !
! D21 D(4,5,12,1) -0.0042 -DE/DX = 0.0 !
! D22 D(4,5,12,11) -180.0033 -DE/DX = 0.0 !
! D23 D(10,5,12,1) 179.9993 -DE/DX = 0.0 !
! D24 D(10,5,12,11) 0.0003 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency Analysis
Job type: Frequency Method: B3LYP Basis set: 6-31G(d,p) Charge:-1
The log file of the frequency analysis:click here
The D space calculation is provided:[| click here]

| Boratabenzene frequency analysis | ||
| File Name | Boratabenzene_opt_fre | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | -1 | |
| Spin | Singlet | |
| E(RB3LYP) | -219.02052984 | a.u. |
| RMS Gradient Norm | 0.00015817 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 11.0032 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 5 minutes 5.0 seconds. | ||
The item table from the "real" output is showing below.
Item Value Threshold Converged? Maximum Force 0.000434 0.000450 YES RMS Force 0.000158 0.000300 YES Maximum Displacement 0.000888 0.001800 YES RMS Displacement 0.000399 0.001200 YES Predicted change in Energy=-7.208081D-07 Optimization completed. -- Stationary point found. GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
low frequencies are showing below:
Low frequencies --- -13.1144 0.0005 0.0009 0.0013 15.0608 18.1770 Low frequencies --- 371.3452 404.2345 565.2524
Population
Job type: Energy Method: B3LYP Basis set: 6-31G(d,p) Additional keywords: pop=full, NBO: Full NBO Charge: -1
The log file of the population analysis:click here
The D space calculation is provided:[| click here]

| Boratabenzene population analysis | ||
| File Name | BORATABENZENE_OPT_PO | |
| File Type | .log | |
| Calculation Type | SP | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | -1 | |
| Spin | Singlet | |
| E(RB3LYP) | -219.02052984 | a.u. |
| RMS Gradient Norm | a.u. | |
| Imaginary Freq | ||
| Dipole Moment | 11.0032 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours minutes 45.0 seconds. | ||
Borazine
Optimisation
Job type: Optimisation Method: B3LYP Basis set: 6-31G(d,p)
The log file of optimisation:click here
The D space calculation is provided:[| click here]
| Borazine optimisation | ||
| File Name | Borazine | |
| File Type | .log | |
| Calculation Type | FOPT | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -242.68459789 | a.u. |
| RMS Gradient Norm | 0.00007128 | a.u. |
| Imaginary Freq | ||
| Dipole Moment | 0.0003 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 4 minutes 33.0 seconds. | ||
The "real" output:the final set of forces and displacements,which gives an indication of the calculation convergence
Item Value Threshold Converged?
Maximum Force 0.000117 0.000450 YES
RMS Force 0.000036 0.000300 YES
Maximum Displacement 0.000327 0.001800 YES
RMS Displacement 0.000104 0.001200 YES
Predicted change in Energy=-1.206132D-07
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,11) 1.0097 -DE/DX = 0.0 !
! R2 R(2,12) 1.1949 -DE/DX = 0.0001 !
! R3 R(3,10) 1.0097 -DE/DX = 0.0 !
! R4 R(4,8) 1.1949 -DE/DX = 0.0001 !
! R5 R(5,9) 1.0097 -DE/DX = 0.0 !
! R6 R(6,7) 1.1949 -DE/DX = 0.0001 !
! R7 R(7,9) 1.4307 -DE/DX = 0.0 !
! R8 R(7,11) 1.4306 -DE/DX = 0.0 !
! R9 R(8,9) 1.4307 -DE/DX = -0.0001 !
! R10 R(8,10) 1.4307 -DE/DX = -0.0001 !
! R11 R(10,12) 1.4306 -DE/DX = 0.0 !
! R12 R(11,12) 1.4307 -DE/DX = -0.0001 !
! A1 A(6,7,9) 121.4339 -DE/DX = 0.0 !
! A2 A(6,7,11) 121.4439 -DE/DX = 0.0 !
! A3 A(9,7,11) 117.1222 -DE/DX = 0.0 !
! A4 A(4,8,9) 121.4457 -DE/DX = 0.0 !
! A5 A(4,8,10) 121.4367 -DE/DX = 0.0 !
! A6 A(9,8,10) 117.1176 -DE/DX = 0.0 !
! A7 A(5,9,7) 118.5554 -DE/DX = 0.0 !
! A8 A(5,9,8) 118.5605 -DE/DX = 0.0 !
! A9 A(7,9,8) 122.884 -DE/DX = 0.0 !
! A10 A(3,10,8) 118.5663 -DE/DX = 0.0 !
! A11 A(3,10,12) 118.5598 -DE/DX = 0.0 !
! A12 A(8,10,12) 122.8739 -DE/DX = 0.0 !
! A13 A(1,11,7) 118.5621 -DE/DX = 0.0 !
! A14 A(1,11,12) 118.5672 -DE/DX = 0.0 !
! A15 A(7,11,12) 122.8707 -DE/DX = 0.0 !
! A16 A(2,12,10) 121.437 -DE/DX = 0.0 !
! A17 A(2,12,11) 121.4314 -DE/DX = 0.0 !
! A18 A(10,12,11) 117.1316 -DE/DX = 0.0 !
! D1 D(6,7,9,5) -0.0006 -DE/DX = 0.0 !
! D2 D(6,7,9,8) 179.9979 -DE/DX = 0.0 !
! D3 D(11,7,9,5) 179.9989 -DE/DX = 0.0 !
! D4 D(11,7,9,8) -0.0026 -DE/DX = 0.0 !
! D5 D(6,7,11,1) 0.0011 -DE/DX = 0.0 !
! D6 D(6,7,11,12) -179.9993 -DE/DX = 0.0 !
! D7 D(9,7,11,1) -179.9984 -DE/DX = 0.0 !
! D8 D(9,7,11,12) 0.0012 -DE/DX = 0.0 !
! D9 D(4,8,9,5) 0.0008 -DE/DX = 0.0 !
! D10 D(4,8,9,7) -179.9977 -DE/DX = 0.0 !
! D11 D(10,8,9,5) 180.0 -DE/DX = 0.0 !
! D12 D(10,8,9,7) 0.0015 -DE/DX = 0.0 !
! D13 D(4,8,10,3) -0.0012 -DE/DX = 0.0 !
! D14 D(4,8,10,12) 180.0002 -DE/DX = 0.0 !
! D15 D(9,8,10,3) 179.9995 -DE/DX = 0.0 !
! D16 D(9,8,10,12) 0.0009 -DE/DX = 0.0 !
! D17 D(3,10,12,2) 0.0011 -DE/DX = 0.0 !
! D18 D(3,10,12,11) -180.0008 -DE/DX = 0.0 !
! D19 D(8,10,12,2) 179.9997 -DE/DX = 0.0 !
! D20 D(8,10,12,11) -0.0022 -DE/DX = 0.0 !
! D21 D(1,11,12,2) -0.0012 -DE/DX = 0.0 !
! D22 D(1,11,12,10) 180.0007 -DE/DX = 0.0 !
! D23 D(7,11,12,2) -180.0008 -DE/DX = 0.0 !
! D24 D(7,11,12,10) 0.0011 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
Frequency Analysis
Job type: Frequency Method: B3LYP Basis set: 6-31G(d,p)
The log file of the frequency analysis:click here
The D space calculation is provided:[| click here]

| Borazine frequency analysis | ||
| File Name | Borazine_opt_fre | |
| File Type | .log | |
| Calculation Type | FREQ | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -242.68459788 | a.u. |
| RMS Gradient Norm | 0.00007138 | a.u. |
| Imaginary Freq | 0 | |
| Dipole Moment | 0.0003 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours 4 minutes 57.0 seconds. | ||
The item table from the "real" output is showing below.
Item Value Threshold Converged?
Maximum Force 0.000206 0.000450 YES
RMS Force 0.000071 0.000300 YES
Maximum Displacement 0.000378 0.001800 YES
RMS Displacement 0.000137 0.001200 YES
Predicted change in Energy=-1.388898D-07
Optimization completed.
-- Stationary point found.
low frequencies are showing below:
Low frequencies --- -11.1723 -0.0003 -0.0001 0.0012 8.7743 11.1553 Low frequencies --- 288.5020 290.4072 404.0137
Population
Job type: Energy Method: B3LYP Basis set: 6-31G(d,p) Additional keywords: pop=full, NBO: Full NBO
The log file of the population analysis:click here
The D space calculation is provided:[| click here]
| Borazine population analysis | ||
| File Name | BORAZINE_OPT_PO | |
| File Type | .log | |
| Calculation Type | SP | |
| Calculation Method | RB3LYP | |
| Basis Set | 6-31G(d,p) | |
| Charge | 0 | |
| Spin | Singlet | |
| E(RB3LYP) | -242.68459789 | a.u. |
| RMS Gradient Norm | a.u. | |
| Imaginary Freq | ||
| Dipole Moment | 0.0003 | Debye |
| Point Group | C1 | |
| Job cpu time: 0 days 0 hours minutes 51.0 seconds. | ||
Compare and contrast
According to the table above, some analysis has been done to discover physical properties of the molecules.These four molecules that we are interested are all aromatic systems. This will lead us to have a better understanding about their structure
Difference in Charge Distribution
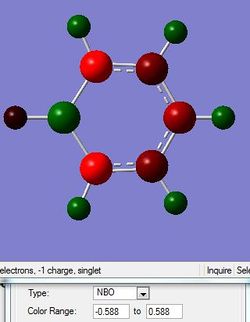
One of the important properties about these aromatic ssysterm is the charge distribution which has been interpreted
| Benzene | Pyridinium | Boratabenzene | Borazine | |
|---|---|---|---|---|
| Charge distribution by colour | 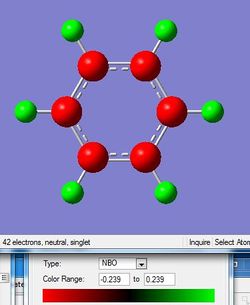
|

|
|

|
| Charge distribution by number | 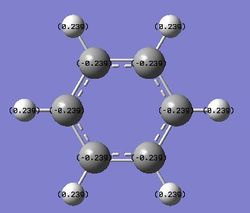
|
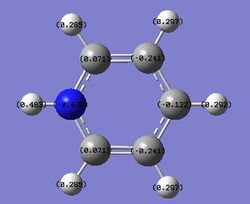
|
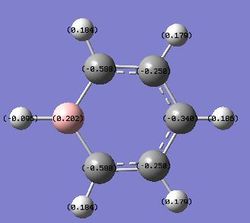
|
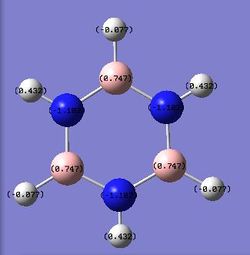
|
For benzene, the six member ring only consists of carbon atom. All the carbon atoms are bonded covalently as there is no difference between two bonded atoms. So the whole system is conjugated and the electrons are completely delocalised over the system. So the charges on carbon atoms are the same,-0.239. Each of the carbon atoms is also singly bonded to a hydrogen atom which is less electronegative than carbon. So the carbon hydrogen bond is not pure covalent and the hydrogen atoms are all slightly positively charged. This explains the reason for the negative value of the charge on the carbon as well as the positive value for the hydrogen.
Pyridine is isoelectronic with benzene. However, one of the carbon atoms is replaced by a nitrogen atom which is more electronegative than carbon. It leads to an electron localisation around the nitrogen atoms on the ring. This gives the nitrogen atom a negative charge,-0.470, and a positive charge, 0.071, on the adjacent carbons. However, this electron localisation does not affect the carbon atoms which are not next to nitrogen.
For borabenzene, the electron localisation effect appears due to the electronegativity difference between boron and carbon atom. In this case, the boron atom has a positive charge while the adjacent carbon atoms have negative charge.
For borazine, the six member ring consists of three boron atoms and three nitrogen atoms. The ring is still aromatic as the lone pair of electrons on the nitrogen is delocalised above and below the ring to form a pi bonding. However, compared with nitrogen atom, the boron is much more electron positive. As a result, the nitrogen is negatively charged while the boron is slightly positive charged, -1.102 and 0.747, respectively.

Difference in molecular orbitals

Molecular orbitals and aromaticity
Aromaticity is the property that is usually referred as a property that the conjugated rings with lone pair of electrons or unsaturated bonds have a stronger stabilisation than that would be expected. It can be interpreted in terms of electron delocalisation and resonance. More specific, the delocalised electrons are one type of electrons that can be shared over two bonding atoms, in other words, they do not locate at a single bond. For aromaticity rings, like benzene, the six p orbital being orthogonal to the planar overlap side by side with each other to form the pi bond. The electrons within this pi bond are delocalised as they are shared over 2 atoms. They are moving a circular motion above and below the ring which inducing a ring current. A well-known theory is introduced at this stage which is the huckel’s rule. In huckel’s rule, for an aromatic system, it always have a pi delocalised electron for 4N+2, in which N can be 0,1,2,3,4…
Comparison of molecular orbitals
For molecular orbital 7, this is a sigma bonding orbital of valence s electrons. The LCAO is a symmetrical planner with a shape of star. For benzene, as the ring consists of only carbon, so the electron is delocalised completely in the pi orbitals. So the benzene calculated MO is exactly the same as LCAO. However, it is not the case for the other three aromatic rings due to the partially electron localisation effect. As nitrogen is more electronegative than carbon and boron is more electropositive than carbon, the MO for pyridinium is pentacle due to the electron localisation on nitrogen. The calculated MO shift to carbon in boratabenzene. For borazine, the MO appears as a triangle as the electron density is partially located on the nitrogen atoms. The energy of the MO(all negative) is: pyridinium< borazine< benzene< boratabenzene.The pyridinium has the best orbital overlap, energy level of its MO is low in energy.
For molecular orbital 21, according to the LCAOs, it is the LUMO which has two node planes perpendicular to each other. The electron density is separated in to four part. Each part is out of phase with each other. Due to the nature of the benzene, the part of MO is identical in size. However, the discrepancy in the electronegativity of nitrogen and carbon, the part of MO having nitrogen will have a larger size in the pyridinium. This happens in the opposite way in boratabenzene. Due to the electron location on the nitrogen atoms, borazine MO appears as a hexagonal. The energy of the MO (all negative apart from boratabenzene) is: pyridinium< borazine< benzene< boratabenzene. The boratabenzene has the worstorbital overlap, energy level of its MO is high in energy.
For molecular orbital 17, according to the LCAOs, it is the pi bonding orbital. So there should be two equal size electron clouds below and above the ring. For benzene, it is a hexagonal. However, the electron cloud moves towards nitrogen in pyridinium as well as in borazine and moves away from boron in boratabenzene.The energy of the MO(all negative) is: pyridinium< borazine< benzene< boratabenzene
As the molecule is more symmetric, it will have more degenerate orbitals. Among these four aromatic ring systems, benzene is the most symmetric ring. Borazine is not as symmetric as benzene but it still have more degenerate orbitals than boratabenzene or pyridinium dose. However, the boratabenzene and pyridinium only have one carbon being replaced by an atom of a different element. This reduces their symmetry largely. The electronegativity of the replaced atoms have a significant effect on orbital overlap in terms of increasing or decreasing orbital energy level.
The substitution and molecular orbitals
The subsitition has a fairly significant influenceon the MO. Electronegative substituents or electropositive substituents lead to a poorly overlap between LICAOs. This will increase the bonding orbital energy and decrease the antibondng orbital energy. The energy gap between HOMO and LUMO is reduced and the reactivity of the molecule is increased meanwhile.