Rep:Mod:AMBS96
I investigated the energetics of the Haber process. To do this, I optimised molecules of NH3, N2 and H2 using Gaussview. I then extracted the molecular information including charge, bond length, bond length and vibration properties and at the dynamic images for all three. We used the energy values to calculate the energy change of the Haber-Bosch process. I then analysed CH3OH and itt molecular orbitals and the charge on the atoms.
Ammonia Molecule
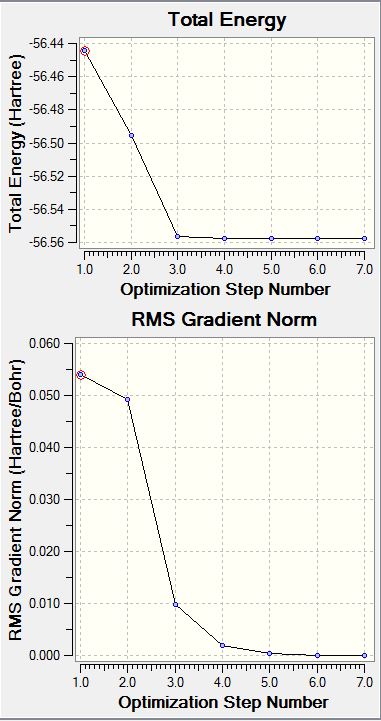
File Name: ASmith_NH3_Molecule_Optimisation File Type: .log Calculation Type: FREQ Calculation Method: RB3LYP Basis Set: 6-31G(d,p) Charge: 0 Spin: Singlet E(RB3LYP): -56.55776873 a.u. RMS Gradient Norm: 0.00000485 a.u. Imaginary Freq: 0 Dipole Moment: 1.8466 Debye Point Group: C3V N-H Bond Length: 1.01798 Å H-N-H Bond Angle: 105.741 degrees
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986265D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
NH3 Optimisation Graphs
NH3 LOG File And jmolApplet
Optimised NH3 Molecule |
File:ASMITH NH3 MOLECULE OPTIMISATION.LOG
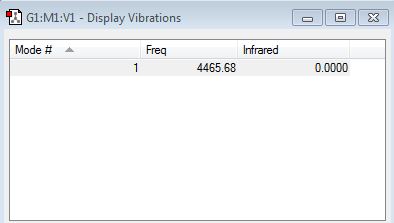
NH3 Display Vibrations
Questions
How many modes do you expect from the 3N-6 rule? 6 - (3x4)-6
Which modes are degenerate (ie have the same energy)? 2&3, 5&6
Which modes are "bending" vibrations and which are "bond stretch" vibrations? Bending: 1, 2, 3 Bond Stretch: 4, 5, 6
Which mode is highly symmetric? 4
One mode is known as the "umbrella" mode, which one is this? 1
How many bands would you expect to see in an experimental spectrum of gaseous ammonia? I would expect to see 2 bands in the experimental spectrum for the 6 vibrational modes of gaseous ammonia due to there being 2 peaks with a high enough relative intensity (Y-axis value) to be seen. The peaks with the low intensity are due to a small change in the dipole moment when the bond vibrates.
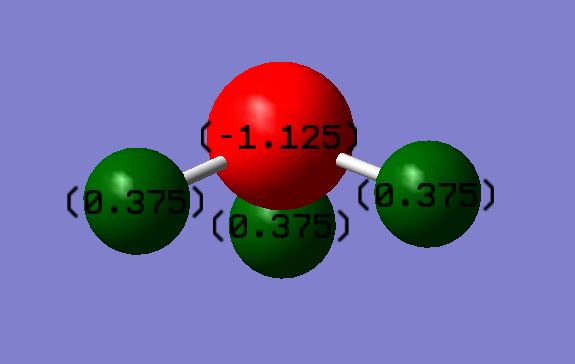
Charge Distribution For Ammonia
Charge On N: -1.125
Charge On H: 0.375
I would expect the Nitrogen to have a negative charge and the Hydrogen to have a positive charge due to Nitrogen having a higher electronegativity.
H2 Molecule
File Name ASMITH_H2_Optimisation File Type .log Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -1.17853936 RMS Gradient Norm 0.00000017 Imaginary Freq 0 Dipole Moment 0.0000 Point Group D*H H2 Bond Length: 0.74279 Å H2 Bond Angle: 180 degrees
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
H2 Optimisation Graphs
H2 LOG File And jmolApplet
Optimised H2 Molecule |
File:ASMITH H2 OPTIMISATION.LOG
H2 Display Vibrations
N2 Molecule
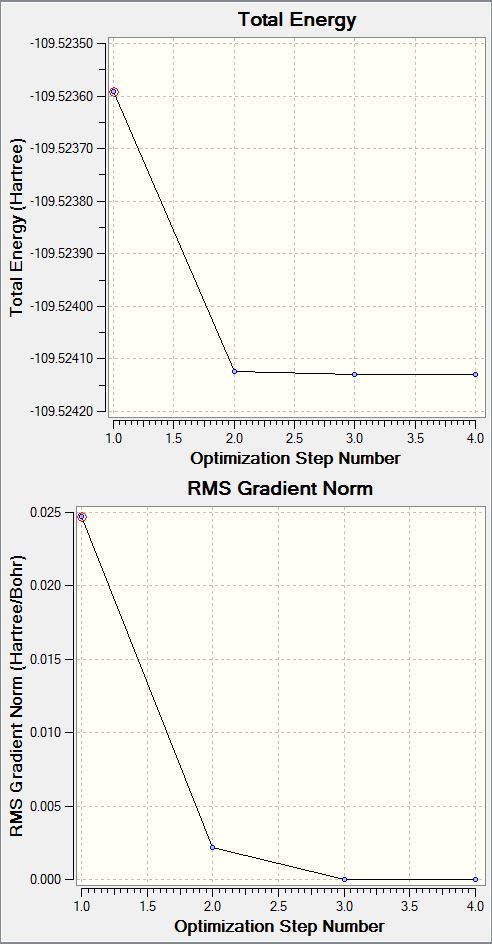
File Name ASMITH_N2_Optimisation File Type .log Calculation Type FREQ Calculation Method RB3LYP Basis Set 6-31G(d,p) Charge 0 Spin Singlet E(RB3LYP) -109.52412868 a.u. RMS Gradient Norm 0.00000060 a.u. Imaginary Freq 0 Dipole Moment 0.0000 Debye Point Group D*H N2 Bond Length: 1.10550 Å N2 Bond Angle: 180 degrees
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401071D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
N2 Optimisation Graphs
N2 LOG File And jmolApplet
Optimised N2 Molecule |
File:ASMITH N2 OPTIMISATION.LOG
N2 Display Vibrations
Haber-Bosch Reaction Energy
N2 + 3h2--> 2NH3
E(NH3)= -56.55776873 a.u.
2*E(NH3)= -56.55776873 a.u.
E(N2)= -109.52412868 a.u.
E(H2)= -1.17853936 a.u.
3*E(H2)= -3.53561808 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579074 a.u.
ΔE= -0.05579074 x 2625.5 = -146.4785879 kJ/mol.
A ΔE value of -146.4785879 kJ/mol indicated that energy is released when H2 and N2 are reacted to form NH3. Therefore Ammonia is more stable than a mixture of H2 and N2 gas.
A literature value[1] gives the enthalpy change of formation of Ammonia to be -9.34 (±0.01) kcal/mol or -39.08 kj/mol. This could be different due to temperature/pressure and solvation effects during the reaction.
CH3OH Molecule
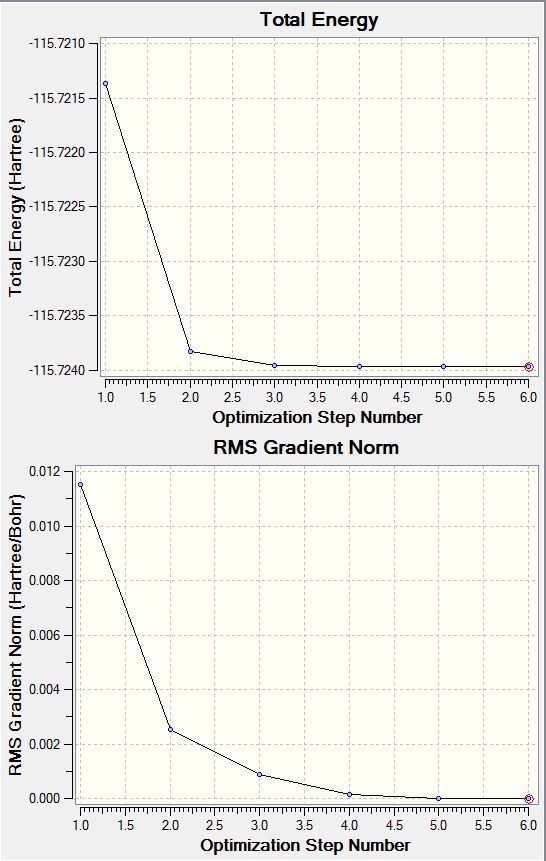
File Name: ASMITH_CH3OH_Optimisation File Type: .log Calculation Type: FREQ Calculation Method: RB3LYP Basis Set: 6-31G(d,p) Charge: 0 Spin: Singlet E(RB3LYP): -115.72396437 a.u. RMS Gradient Norm: 0.00001494 a.u. Imaginary Freq: 0 Dipole Moment: 1.6635 Debye Point Group: C1
Item Value Threshold Converged?
Maximum Force 0.000038 0.000450 YES
RMS Force 0.000020 0.000300 YES
Maximum Displacement 0.000338 0.001800 YES
RMS Displacement 0.000153 0.001200 YES
Predicted change in Energy=-1.480106D-08
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1006 -DE/DX = 0.0 !
! R2 R(1,3) 1.093 -DE/DX = 0.0 !
! R3 R(1,4) 1.1006 -DE/DX = 0.0 !
! R4 R(1,5) 1.4181 -DE/DX = 0.0 !
! R5 R(5,6) 0.9652 -DE/DX = 0.0 !
! A1 A(2,1,3) 107.8992 -DE/DX = 0.0 !
! A2 A(2,1,4) 108.2583 -DE/DX = 0.0 !
! A3 A(2,1,5) 112.8281 -DE/DX = 0.0 !
! A4 A(3,1,4) 107.8997 -DE/DX = 0.0 !
! A5 A(3,1,5) 106.9041 -DE/DX = 0.0 !
! A6 A(4,1,5) 112.8274 -DE/DX = 0.0 !
! A7 A(1,5,6) 107.7375 -DE/DX = 0.0 !
! D1 D(2,1,5,6) 61.5104 -DE/DX = 0.0 !
! D2 D(3,1,5,6) 179.9639 -DE/DX = 0.0 !
! D3 D(4,1,5,6) -61.5824 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
GradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGradGrad
CH3OH Optimisation Graphs
H2 LOG File And jmolApplet
Optimised H2 Molecule |
File:ASMITH CH3OH OPTIMISATION.LOG
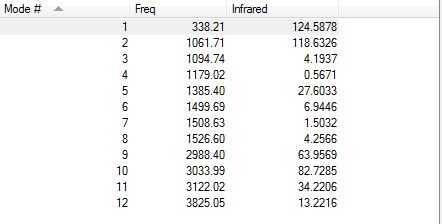
CH3OH Display Vibrations
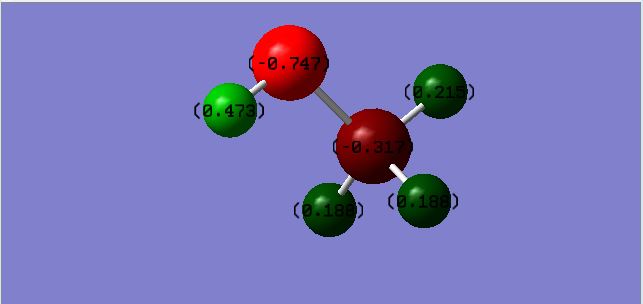
Charge Distribution For CH3OH
Charge On C: -0.317
Charge On O: -0.747
Charge On CH: 0.188,0.188,0.215 - One of the hydrogens has a different value compared to the other 2 due to the hydrogen with the higher charge being closer to the Oxygen. The other 2 hydrogens are in equivalent positions and therefore have the same charge.
Charge On OH: 0.473 - The charge on this hydrogen is higher than the hydrogens attached to the carbon atom due to the Oxygen having a higher electronegativity.
CH3OH MO's
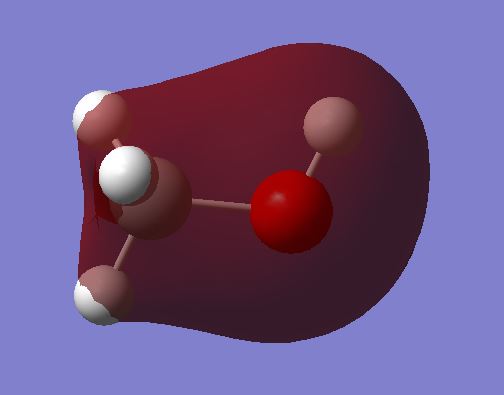 MO 3 shows the bonding molecular orbital as a result of the overlap of:
2s AO overlap between C and O. The MO is occupied.
MO 3 shows the bonding molecular orbital as a result of the overlap of:
2s AO overlap between C and O. The MO is occupied.
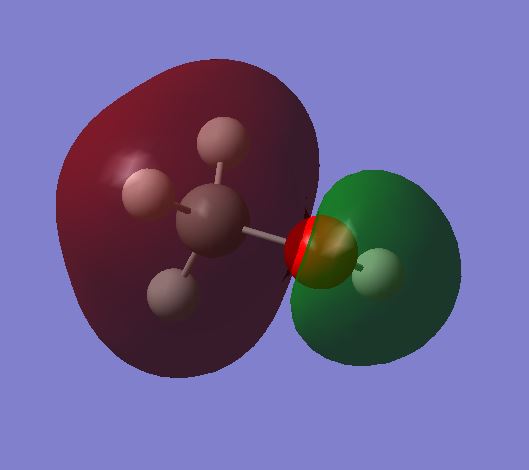 MO 4 shows the antibonding molecular orbital of MO 3. This is due to:
The overlap of the 2s orbital on the C and the 2s orbital on the O. This gives an antibonding MO due to the 2s orbitals being out of phase. The MO is occupied.
The orbital plot shows a large amount of electron density around the Carbon. This is strange as this would be expected for the oxygen instead due to it having a higher electronegativity. The larger MO plot on the carbon could be due to the bonds between the Carbon and the Hydrogens which would give an area of high electron density.
MO 4 shows the antibonding molecular orbital of MO 3. This is due to:
The overlap of the 2s orbital on the C and the 2s orbital on the O. This gives an antibonding MO due to the 2s orbitals being out of phase. The MO is occupied.
The orbital plot shows a large amount of electron density around the Carbon. This is strange as this would be expected for the oxygen instead due to it having a higher electronegativity. The larger MO plot on the carbon could be due to the bonds between the Carbon and the Hydrogens which would give an area of high electron density.
 MO 5 shows the bonding molecular orbital as a result of the overlap of:
Overlap of 2py AO on the C and 2py AO on the O. The MO is occupied.
MO 5 shows the bonding molecular orbital as a result of the overlap of:
Overlap of 2py AO on the C and 2py AO on the O. The MO is occupied.
 MO 8 shows the antibonding molecular orbital of MO 5 as a result of the overlap of:
Overlap of 2py AO on the C and 2py AO on O which are out of phase. The MO is occupied.
MO 8 shows the antibonding molecular orbital of MO 5 as a result of the overlap of:
Overlap of 2py AO on the C and 2py AO on O which are out of phase. The MO is occupied.
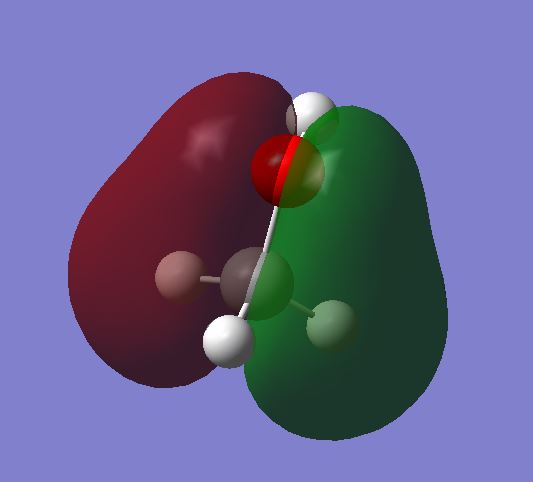 MO 6 shows the bonding molecular orbital as a result of the overlap of:
2pz AO on the C and the 2pz AO on the O. The MO is occupied.
MO 6 shows the bonding molecular orbital as a result of the overlap of:
2pz AO on the C and the 2pz AO on the O. The MO is occupied.
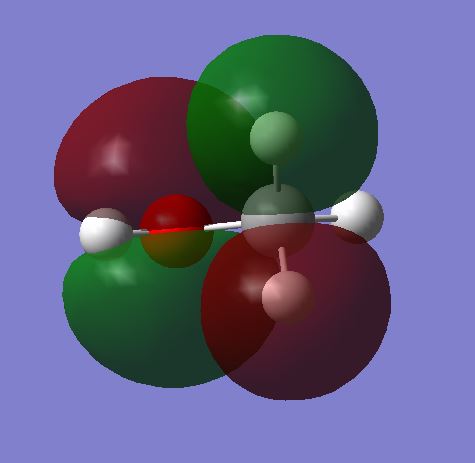 MO 9 shows the antibonding molecular orbital of 6 as a result of the overlap of:
2pz AO on the C and the 2pz AO on the O which are out of phase with each other. This is the HOMO.
MO 9 shows the antibonding molecular orbital of 6 as a result of the overlap of:
2pz AO on the C and the 2pz AO on the O which are out of phase with each other. This is the HOMO.
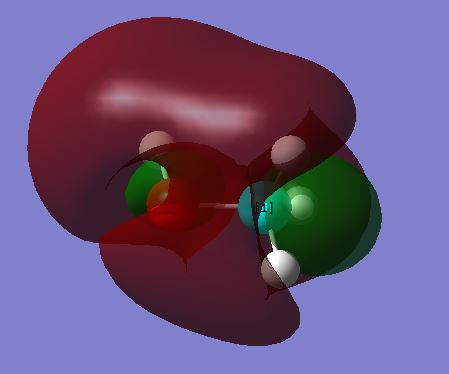 This is the LUMO. The plot looks strange and is hard to interpret.
This is the LUMO. The plot looks strange and is hard to interpret.
References
- ↑ G A Petersson John Mantzaris Journal of the American Chemical Society. , 1991, Vol.113(7), p.2486-2490