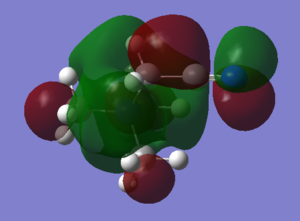
Rep:Mod:ALURBAS
Module 2
BH3 Molecule
Using GaussView 5.0 a BH3 was created, the bond lengths were then adjusted to 1.5 angstrom. Optimisation of the molecule was preformed using "Gaussian calculation setup," with Job Type as optimisation, B3LYP as the method and finally 3-21G as the basis set. The optimisation of BH3 gives us the lowest energy positions of all four of the nuclei in the molecule, the method tells the program what approximations that are being used when solving the Schrodinger equation and the basis set determines the accuracy (3-21G basis set has very low accuracy but it allows fast computation of the results).
Optimisation produced a BH3 molecule with a B-H bond distance of 1.19349 angstrom and a H-B-H angle of 1200. A summary of the BH3 molecule optimisation shown below.

But has the molecule been optimised? It looks very similar to the created BH3, to give evidence of the optimisation we look to the summary, shown right, giving a gradient close to zero gives some proof but checking the .log file as seeing if it has converged makes it definitive. The .log file indicates all the information produced during the Gaussian calculation, near the end of the log file (attached below) a table is seen indicating that the convergence has occurred.
Item Value Threshold Converged?
Maximum Force 0.000413 0.000450 YES
RMS Force 0.000271 0.000300 YES
Maximum Displacement 0.001610 0.001800 YES
RMS Displacement 0.001054 0.001200 YES
Predicted change in Energy=-1.071764D-06
Optimization completed.
-- Stationary point found.
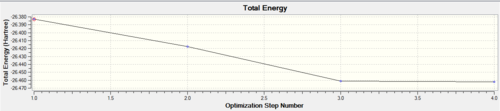
Opening the log file with the "Read Intermediate Geometries" allows additional graphs to be viewed (shown below), showing the change in energy and gradient as the optimisation takes place. The Total Energy graph shows each optimisation step going lower in energy until it reaches the optimised structure, the second graph shows Root Mean Squared Gradient graph and as each optimisation occurs it becomes closer to zero. When almost zero gradient obtained the program knows that the optimised structure has been achieved.
A Better Basis Set
The 3-21G while limited by its accuracy provides a good baseline that can be improved upon with better basis sets. The 3-21G LOG file, seen above, is optimised again using 6-31G(d,p) as the basis set, with a higher degree of accuracy. This improved optimisation gave a B-H bond distance of 1.19349 angstrom and a H-B-H angle of 1200. An image of the optimised BH3 shown below.
The following table confirms optimisation of the molecule, similar to the 3-21G basis set.

Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000003 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000012 0.001200 YES
Predicted change in Energy=-1.304899D-10
Optimization completed.
-- Stationary point found.
File:Georgedoucy BH3 OPT 6-31G DP.LOG
The total energy for the 3-21G optimised molecule was -26.46226338 au and for the 6-31G -26.61532181 au, this gives a difference of 0.15305843 au. The difference may not seem like much but when converted to kj/mol (0.15305843*2625.50) gives 402 kj/mol a huge difference in energy this gives reason to why two different basis sets are never compared to each other and why energies in au are reported so accurately (7 dp).

Frequency Analysis
Using the 6-31G(d,p) optimised structure complete, frequency analysis can now be preformed. Frequency analysis tells us if the molecule is in a maxima, a transition state, or a minima, the ground state, and also gives us the IR and Raman modes. If all the frequencies obtained are positive the molecule is in a ground state and if one is negative it is in a transition state, the analysis is run on the 6-31G(d,p) optimisation with the job type changed to frequency; summary and tables shown below (with proof of optimisation).

Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000019 0.001800 YES
RMS Displacement 0.000009 0.001200 YES
Predicted change in Energy=-1.323374D-10
Optimization completed.
-- Stationary point found.
Low frequencies --- -0.9033 -0.7343 -0.0054 6.7375 12.2491 12.2824
Low frequencies --- 1163.0003 1213.1853 1213.1880
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A2" E' E'
Frequencies -- 1163.0003 1213.1853 1213.1880
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9986 0.9601 0.9601
IR Inten -- 92.5478 14.0553 14.0589
Atom AN X Y Z X Y Z X Y Z
1 5 0.00 0.00 0.16 0.00 0.10 0.00 -0.10 0.00 0.00
2 1 0.00 0.00 -0.57 0.00 0.08 0.00 0.81 0.00 0.00
3 1 0.00 0.00 -0.57 -0.39 -0.59 0.00 0.14 0.39 0.00
4 1 0.00 0.00 -0.57 0.39 -0.59 0.00 0.14

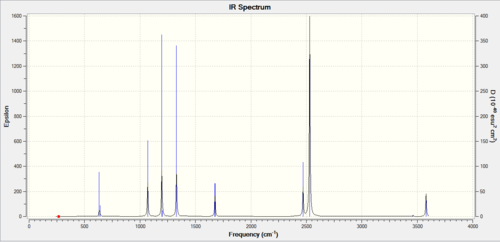
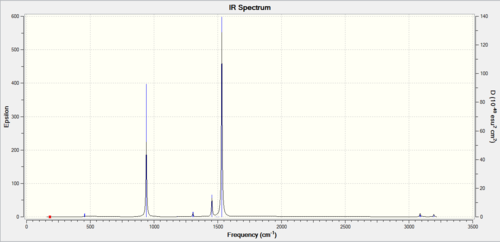
The Low frequency line gives us the proof we needed of correct optimisation, with them being close to zero and orders of magnitude lower than the the real frequencies (12.2824 to 1231.1880). The closer the low frequencies are to zero the more accurate the calculation. The stationary point has been found now a look at the vibrations of the molecule.

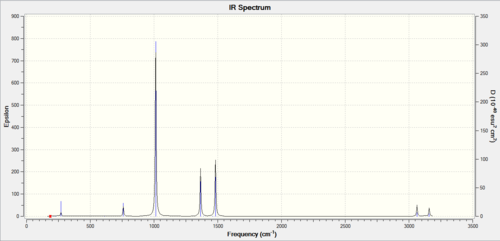
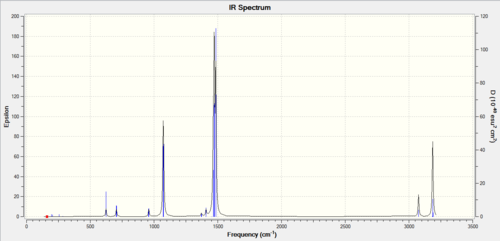
The three peaks seen in the spectrum above are 1163, 1213, 2715 as they cause dipole moments which are the only peaks that show up on IR, the other peaks do not show up as they do not have an overall dipole moments in the molecule.
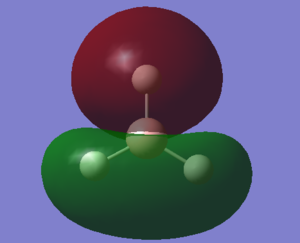
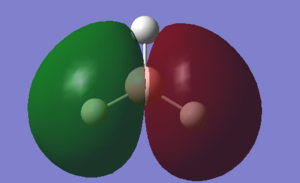
Molecular Orbitals
With the above calculations leading to an optimised BH3 molecule it also allows us to visualise molecular orbitals using the .chk file, this file was submitted to SCAN and job type changed to energy, keywords pop=full and NBO turned fully on. The D-space file and the .chk file shown below.
DOI:10042/to-http://hdl.handle.net/10042/23459
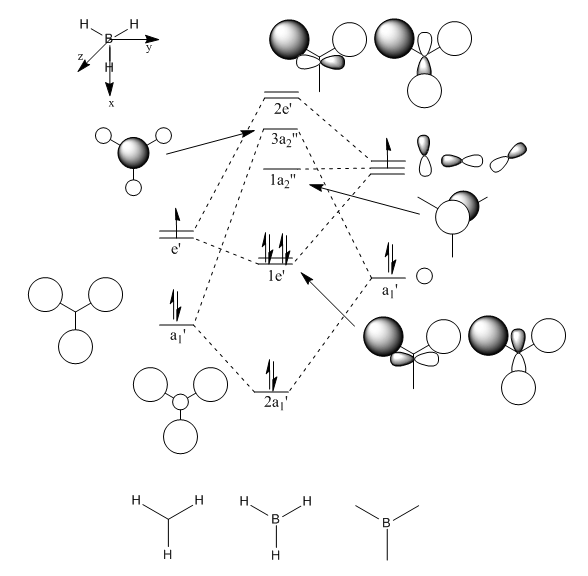
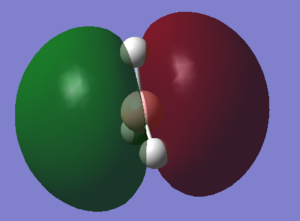
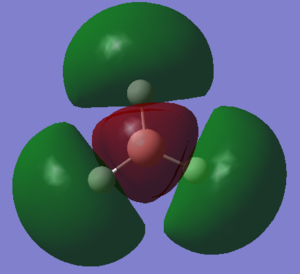
With a Linear Combination of Atomic Orbitals (LCAO) diagram for BH3 as reference the molecular orbitals visualised in Gaussview can be compared and in turn the accuracy of the calculations can be reviewed.
The above table shows how similar the visualised to LCAO phase patterns can be but as it goes down to the more diffuse, large molecules some errors can be seen (no. 7) indicating while comparison is good it is not without its faults.
TlBr3
Optimisation
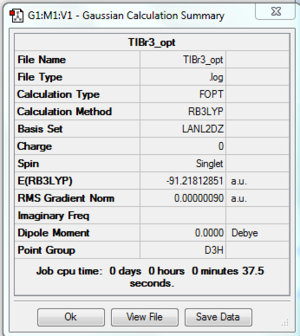
Moving on from the small BH3 molecule the much larger TiBr3 is created, with a total of 186 electrons this requires the use of more advanced processing power to optimised the molecule. Again, using GaussView 5.0 the TlBr3 molecule was made its geometry was tightly restricted to D3h and the calulation set to optimisation using B3LYP and a medium basis set of LanL2DZ this job was submitted to SCAN and the files are shown below. The Tl-Br bond distance was found to be 2.65095 angstrom, the lit value of Tl-Br was found to be 2.512 angstrom, and the Br-Tl-Br bond angle to be 1200. The proof that the molecule was fully optimised shown below. File:Georgedoucy TlBr3 opt.log
DOI:10042/to- http://hdl.handle.net/10042/23495
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000014 0.001200 YES
Predicted change in Energy=-6.084027D-11
Optimization completed.
-- Stationary point found.
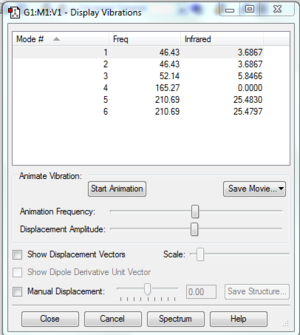
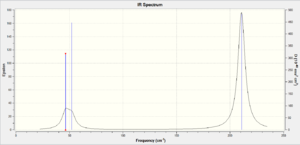
Frequency Analysis
Frequency analysis also preformed on SCAN, convergence proof below with low frequency from .log file. File:Georgedoucy TIBr3 freq.log DOI:10042/to-http://hdl.handle.net/10042/23628
Item Value Threshold Converged?
Maximum Force 0.000002 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000022 0.001800 YES
RMS Displacement 0.000011 0.001200 YES
Predicted change in Energy=-5.660901D-11
Optimization completed.
-- Stationary point found.
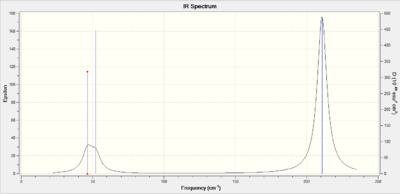
Like the BH3 frequency analysis the six low frequencies are shown to be close to zero indicating a ground state has been found.
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367
Low frequencies --- 46.4289 46.4292 52.1449
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
E' E' A2"
Frequencies -- 46.4289 46.4292 52.1449
Red. masses -- 88.4613 88.4613 117.7209
Frc consts -- 0.1124 0.1124 0.1886
IR Inten -- 3.6867 3.6867 5.8466
Atom AN X Y Z X Y Z X Y Z
1 81 0.00 0.28 0.00 -0.28 0.00 0.00 0.00 0.00 0.55
2 35 0.00 0.26 0.00 0.74 0.00 0.00 0.00 0.00 -0.48
3 35 0.43 -0.49 0.00 -0.01 -0.43 0.00 0.00 0.00 -0.48
4 35 -0.43 -0.49 0.00 -0.01 0.43 0.00 0.00 0.00 -0.48

BBr3
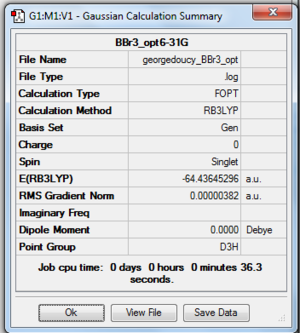
In this experiment run, the heavy bromide atoms requires a pseudo-potential as the Schrodinger equation can not deal with these heavy atoms. The pseudo-potential allows the core of heavy atom to be modeled, the boron atom can be calculated with a normal basis set. Using the 6-31G(d,p) BH3 optimised log file, the hydrogens are changed to bromine atoms in calulation setup the method is changed to GEN with additional keyword of "pseudo=read gfinput. This allows the pseudo potential for each individual atom to be set, with boron using 6-31G(d,p) and the bromine using LanL2DZ (this is done by going into the file and editing it manually) the job is than submitted to SCAN, files shown below.
After the optimisation the B-Br bond distance was found to be 1.93396 angstrom with a bond angle of 1200, the proof of optimisation shown below. DOI:10042/to-http://hdl.handle.net/10042/23454
Item Value Threshold Converged?
Maximum Force 0.000008 0.000450 YES
RMS Force 0.000005 0.000300 YES
Maximum Displacement 0.000036 0.001800 YES
RMS Displacement 0.000023 0.001200 YES
Predicted change in Energy=-4.027544D-10
Optimization completed.
-- Stationary point found.
Comparison and Discussion
| Molecule | Distance/angstrom |
|---|---|
| BH3 | 1.19 |
| BBr3 | 1.93 |
| TlBr3 | 2.65 |
BH3 and BBr3 are the easiest to compare as the only substitution is hydrogen for bromine, they are both lewis acids with incomplete octets. However, the lone pairs on the bromine atoms can donate to the electron deficient boron atom leading to the lewis acidity of BBr3 to be lower than BH3 with it having a shorter bond distance. Bromine also being a much larger and more diffuse atom also contributes to it having a longer bond distance that BH3.
Comparing the central atoms of BBr3 and TlBr3, boron and thallium are both group 13 atoms with thallium being a heavier atom with access to d electrons however due to the inert pair effect Tl more likely to be in oxidation step (I). The size of the thallium atom is the major contributor to the difference in bond distance, the mass also contributes to the differences seen in the vibrational spectra.
Comparison of Vibrational Spectra
| Molecule | Frequency/cm-1 |
|---|---|
| BH3 | 1163.00, 1213.19, 1213.19, 2582.26, 2715.43, 2715.43 |
| TlBr3 | 46.43, 46.43, 52.14, 165.27, 210.69, 210.69 |
The BH3, left spectra, and the TlBr3, the right spectra above, are both have there similarities and differences. They both have three peaks, with the thallium seeming to join in to one broad peak, the differences in frequencies seen is majorly the large thallium atom but the peaks follow each other closely.
Discussion
This experiment gives insight of the power of computational chemistry as TlBr3 is highly toxic but this approach allows comparisons with different molecules and insight of its bonding.
In the BH3 optimisation when "Reactive Intermediate Geometries" was on Gaussview pictured the molecule with what looked like no bonds. Gaussview defines bond by a set distance but if the distance goes beyond this set amount there is no bond seen between the molecule however that does not mean the bond is gone.
The same method and basis set is needed to compare the optimisation or frequency analysis, as shown by the massive difference in energy between the 3-21G and the 6-31G(d,p) indicates the need for consistent method and basis set to be able to compare fairly. The need for frequency analysis is because it is the second derivative of the potential energy surface (PES) if positive the molecule is at a minimum and if at a negative the molecule is in a transition state and not fully optimised, the IR spectra is also very useful.C
NH3
Optimisation
Using optimisation as the job type, with B3LYP as the method and 6-31G(d,p) the basis set, 6-31G(d,p) used as the molecule is very small giving more accuracy without sacrificing time, additional keywords were "nosymm". The files and proof of optimisation shown below. File:GEORGEDOUCY NH3 OPT.LOG

Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000012 0.000300 YES
Maximum Displacement 0.000079 0.001800 YES
RMS Displacement 0.000053 0.001200 YES
Predicted change in Energy=-1.629731D-09
Optimization completed.
-- Stationary point found.
Frequency Analysis
File and images below detail the proof of optimisation to the ground state. File:GEORGEDOUCY NH3 FREQ.LOG

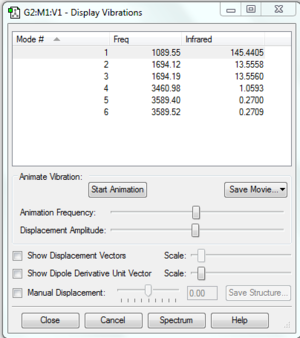
Item Value Threshold Converged?
Maximum Force 0.000021 0.000450 YES
RMS Force 0.000009 0.000300 YES
Maximum Displacement 0.000078 0.001800 YES
RMS Displacement 0.000039 0.001200 YES
Predicted change in Energy=-1.611690D-09
Optimization completed.
-- Stationary point found.
Lowest vibrational frequencies 28 cm-1 which is orders of magnitude from the frequencies labeled, no negative frequencies molecule optimised.
Low frequencies --- -30.7295 -0.0007 0.0008 0.0013 20.1705 28.2664
Low frequencies --- 1089.5535 1694.1244 1694.1856
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 1089.5535 1694.1244 1694.1856
Red. masses -- 1.1800 1.0644 1.0644
Frc consts -- 0.8253 1.8000 1.8001
IR Inten -- 145.4405 13.5558 13.5560
Atom AN X Y Z X Y Z X Y Z
1 7 0.12 0.00 0.00 0.00 -0.02 -0.06 0.00 0.06 -0.02
2 1 -0.53 -0.21 0.00 -0.07 -0.04 0.73 0.25 0.14 0.20
3 1 -0.53 0.11 0.18 0.25 -0.24 -0.03 -0.07 -0.62 0.40
4 1 -0.53 0.11 -0.18 -0.18 0.52 0.18 -0.18 -0.41 -0.36
Molecular Orbitals
Population analysis also preformed with files and MO's shown below. Using the 6-31G optimised .chk file, job type changed to Energy and NBO selected to full NBO, finally key word pop=full. DOI:/10042-to http://hdl.handle.net/10042/23811
| No. | Visualised MO | |
|---|---|---|
| 1 | 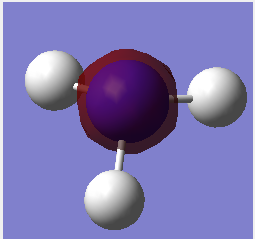
| |
| 2 | 
| |
| 3 | 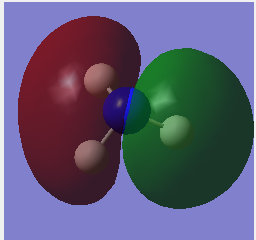
| |
| 4 | 
| |
| 5 | 
| |
| 6 | 
| |
| 7 | 
| |
| 8 | 
|
NBO
Using the population analysis log file, not .chk, the charge distribution of the molecule can be shown(files below). DOI:/10042-to http://hdl.handle.net/10042/23811
Using the "Charge Distribution" option in the results tab, the molecule can be coloured or labeled by its charge but for a quantitative view of the molecule the view file is used, tables below.
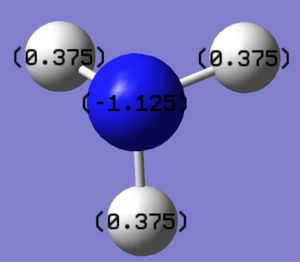
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -1.12515 1.99982 6.11104 0.01429 8.12515
H 2 0.37505 0.00000 0.62250 0.00246 0.62495
H 3 0.37505 0.00000 0.62250 0.00246 0.62495
H 4 0.37505 0.00000 0.62249 0.00246 0.62495
=======================================================================
* Total * 0.00000 1.99982 7.97852 0.02166 10.00000
The above table shows the charge distribution in the molecule, "Natural Charge".
The below table defines that NH3 is a sp3 hybridised molecule, with the first bond in the list showing 69% on the nitrogen atom and 31% contribution on the hydrogen, showing 25% on the s orbital and 75% on the nitrogen. The 4th and 5th item on the list show the atomic nitrogen orbital (1s) and the lone pair in the molecule respectively, also showing sp3 character.
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99909) BD ( 1) N 1 - H 2
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
-0.0001 -0.4986 -0.0059 0.0000 -0.2910
0.0052 0.8155 0.0277 0.0000 0.0000
0.0281 0.0000 0.0000 0.0032 0.0082
( 31.17%) 0.5583* H 2 s( 99.91%)p 0.00( 0.09%)
-0.9996 0.0000 0.0072 -0.0289 0.0000
2. (1.99909) BD ( 1) N 1 - H 3
( 68.83%) 0.8297* N 1 s( 24.86%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2910
-0.0052 0.4077 0.0138 0.7062 0.0240
0.0140 0.0243 0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 3 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 -0.0250
3. (1.99909) BD ( 1) N 1 - H 4
( 68.83%) 0.8297* N 1 s( 24.87%)p 3.02( 75.05%)d 0.00( 0.09%)
0.0001 0.4986 0.0059 0.0000 0.2909
-0.0052 0.4077 0.0138 -0.7062 -0.0239
0.0140 -0.0243 -0.0076 0.0033 0.0031
( 31.17%) 0.5583* H 4 s( 99.91%)p 0.00( 0.09%)
0.9996 0.0000 -0.0072 -0.0145 0.0250
4. (1.99982) CR ( 1) N 1 s(100.00%)
1.0000 -0.0002 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000 0.0000 0.0000 0.0000
5. (1.99721) LP ( 1) N 1 s( 25.38%)p 2.94( 74.52%)d 0.00( 0.10%)
0.0001 0.5036 -0.0120 0.0000 -0.8618
0.0505 0.0000 0.0000 0.0000 0.0000
0.0000 0.0000
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
The table above does not give much information for NH3 but outlines the interactions between molecules, MO mixing, in E(2) anything greater than 20 kcal/mol is of interest.
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (H3N)
1. BD ( 1) N 1 - H 2 1.99909 -0.60417
2. BD ( 1) N 1 - H 3 1.99909 -0.60417
3. BD ( 1) N 1 - H 4 1.99909 -0.60416
4. CR ( 1) N 1 1.99982 -14.16768
5. LP ( 1) N 1 1.99721 -0.31756 24
Finally, this table shows the energy, population or occupation of the bonds in the molecule, including the lone pair. The N-H bonds can all be seen with the same energy with the lone pair being high in energy and the core very low.
NH3BH3
Optimisation
NH3BH3 is made slightly differently in Gaussview first using a ethyl fragment group and then changing the carbons for the needed boron and nitrogen, this molecule needed to be optimised using B3LYP/6-31G(d,p) as it is to be compared with the single BH3 and NH3 molecules and to compare they must have the same basis set and method. Files and proof of optimisation below.
File:Georgedoucy NH3BH3 OPT.LOG

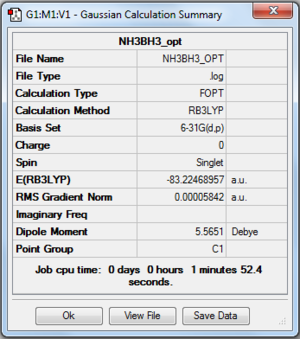
Item Value Threshold Converged?
Maximum Force 0.000124 0.000450 YES
RMS Force 0.000057 0.000300 YES
Maximum Displacement 0.000660 0.001800 YES
RMS Displacement 0.000304 0.001200 YES
Predicted change in Energy=-1.649843D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
Frequency analysis carried out identically as shown before, files and proof of optimisation below.
File:Georgedoucy NH3BH3 FREQ.LOG
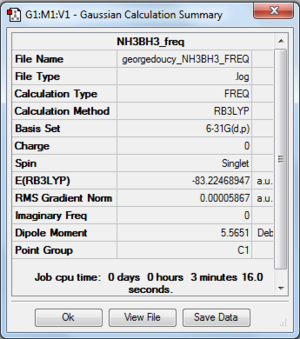
Item Value Threshold Converged?
Maximum Force 0.000112 0.000450 YES
RMS Force 0.000059 0.000300 YES
Maximum Displacement 0.000666 0.001800 YES
RMS Displacement 0.000394 0.001200 YES
Predicted change in Energy=-1.734401D-07
Optimization completed.
-- Stationary point found.
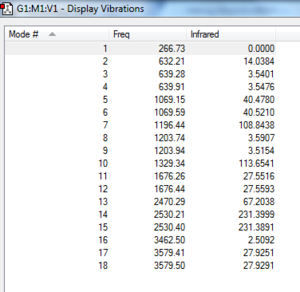
No negative vibrational frequencies, lowest low frequency order of magnitude from vibrational frequency.
Low frequencies --- -0.0014 -0.0010 -0.0004 16.2346 19.0584 42.0053
Low frequencies --- 266.7333 632.2084 639.2804
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 266.7315 632.2083 639.2803
Red. masses -- 1.0078 5.0012 1.0452
Frc consts -- 0.0422 1.1777 0.2517
IR Inten -- 0.0000 14.0384 3.5401
Atom AN X Y Z X Y Z X Y Z
1 1 -0.07 -0.35 0.04 -0.04 0.04 0.28 0.10 0.08 0.42
2 1 0.34 0.11 -0.01 0.01 0.00 0.29 0.15 0.03 -0.06
3 1 -0.27 0.24 -0.03 0.02 0.06 0.28 0.13 -0.04 -0.36
4 1 -0.09 -0.44 0.05 0.00 -0.04 -0.36 0.17 0.11 0.54
5 1 -0.34 0.30 -0.04 0.01 -0.04 -0.36 0.19 -0.03 -0.46
6 1 0.42 0.14 -0.01 0.01 -0.04 -0.36 0.21 0.04 -0.08
7 5 0.00 0.00 0.00 -0.01 0.05 0.48 -0.03 -0.01 0.00
8 7 0.00 0.00 0.00 0.00 -0.04 -0.36 -0.05 -0.01 0.00
4 5 6
Energy Comparison
With NH3BH3 being a joining of BH3 and NH3, molecules that have been optimised already, the energy values of these molecules can be used to calculate the association energy of the combination of the molecules.
E(NH3)= -56.55777 au E(BH3)= -26.61532 au E(NH3BH3)= -83.22469 au
ΔE=E(NH3BH3)-[E(NH3)+(BH3)]
ΔE= -83.22469 au - ( -56.55777 au + -26.61532 au) = -83.22469 au - ( -83.17309 au) = -0.0516 au = -135.48 kj/mol
Mini Project: Ionic Liquids, Designer Solvents
Part one: Comparison of Selected 'Onium' Cations
[N(CH3)4]+
Optimisation
This is the first ionic molecule to be analysed before finally compared and contrasted with the others in this mini-project. In Gaussview 5.0 the [N(CH3)4]+ ion was made by first using a carbon tetrahedral group to make tert-butyl and then changing the centre atom to a nitrogen, the molecule was then optimised using job type optimise, method B3LYP and basis set 6-31G(d,p) and submitted to SCAN (files below), it is important to remember to set the charge to 1 in the method tab.
DOI 10042\to- http://hdl.handle.net/10042/23837
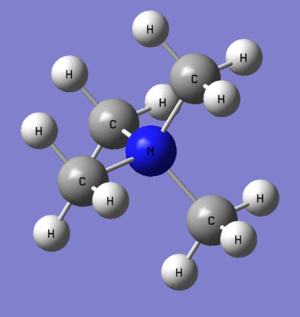
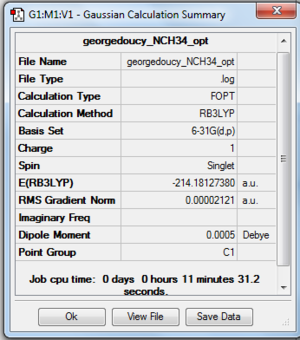
Optimisation gave a tetrahedral ion with a N-C bond distance of 1.50942 angstrom and a C-N-C bond angle of 109.4760, proof of ground state achieved below:
Item Value Threshold Converged?
Maximum Force 0.000052 0.000450 YES
RMS Force 0.000014 0.000300 YES
Maximum Displacement 0.001281 0.001800 YES
RMS Displacement 0.000327 0.001200 YES
Predicted change in Energy=-1.300038D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
[N(CH3)4]+ gave low frequencies that were orders of magnitude smaller than the lowest vibrational frequency. DOI: 10042\to-http://hdl.handle.net/10042/23840
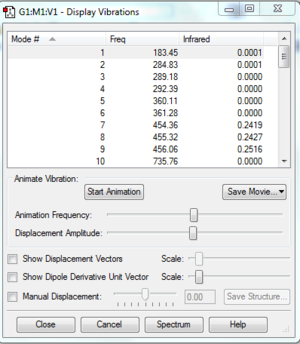
Low frequencies --- -13.4550 -11.5366 0.0009 0.0010 0.0010 12.6011
Low frequencies --- 183.4466 284.8457 289.1865
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 183.4456 284.8341 289.1790
Red. masses -- 1.0079 1.0331 1.0332
Frc consts -- 0.0200 0.0494 0.0509
IR Inten -- 0.0001 0.0001 0.0000
Molecular Orbitals and NBO
Using the .chk file from the optimisation, job type changed to energy, NBO to fully on and keyword pop=full. DOI:10042\to-http://hdl.handle.net/10042/23839 .log file used for NBO and .chk file used for Molecular orbital visualisation, images of charge distribution shown below.
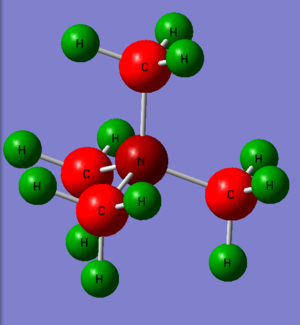
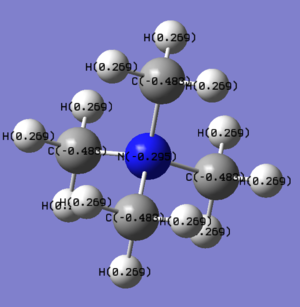
Summary table for charge distribution of every single atom.
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
N 1 -0.29507 1.99950 5.28966 0.00591 7.29507
C 2 -0.48345 1.99947 4.46949 0.01450 6.48345
H 3 0.26906 0.00000 0.72994 0.00100 0.73094
H 4 0.26906 0.00000 0.72993 0.00100 0.73094
H 5 0.26908 0.00000 0.72992 0.00100 0.73092
C 6 -0.48337 1.99947 4.46941 0.01449 6.48337
H 7 0.26905 0.00000 0.72995 0.00100 0.73095
H 8 0.26904 0.00000 0.72996 0.00100 0.73096
H 9 0.26906 0.00000 0.72994 0.00100 0.73094
C 10 -0.48338 1.99947 4.46942 0.01449 6.48338
H 11 0.26904 0.00000 0.72996 0.00100 0.73096
H 12 0.26907 0.00000 0.72993 0.00100 0.73093
H 13 0.26906 0.00000 0.72994 0.00100 0.73094
C 14 -0.48344 1.99947 4.46948 0.01450 6.48344
H 15 0.26907 0.00000 0.72993 0.00100 0.73093
H 16 0.26905 0.00000 0.72995 0.00100 0.73095
H 17 0.26908 0.00000 0.72992 0.00100 0.73092
=======================================================================
* Total * 1.00000 9.99736 31.92671 0.07592 42.00000
The bond contribution between the C-N bonds can be seen to be around 2/3 on the nitrogen and 1/3 on the carbon. 25% s character and 75& p character on the nitrogen indicating a sp3 molecule
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98452) BD ( 1) N 1 - C 2
( 66.35%) 0.8145* N 1 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 -0.5000 0.0007 0.0000 0.8283
-0.0001 -0.0498 0.0000 0.2473 0.0000
0.0017 -0.0084 0.0005 -0.0141 0.0067
( 33.65%) 0.5801* C 2 s( 20.78%)p 3.81( 79.06%)d 0.01( 0.16%)
-0.0003 -0.4552 0.0237 -0.0026 -0.8498
-0.0361 0.0512 0.0022 -0.2537 -0.0108
0.0039 -0.0192 0.0012 -0.0321 0.0153
2. (1.98452) BD ( 1) N 1 - C 6
( 66.35%) 0.8146* N 1 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 0.5000 -0.0007 0.0000 0.0408
0.0000 -0.2863 0.0000 0.8161 -0.0001
-0.0005 0.0014 -0.0096 -0.0017 0.0148
( 33.65%) 0.5801* C 6 s( 20.77%)p 3.81( 79.06%)d 0.01( 0.16%)
0.0003 0.4551 -0.0237 0.0026 -0.0420
-0.0018 0.2937 0.0125 -0.8374 -0.0355
-0.0011 0.0031 -0.0219 -0.0038 0.0338
3. (1.98452) BD ( 1) N 1 - C 10
( 66.35%) 0.8146* N 1 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 0.5000 -0.0007 0.0000 0.3636
-0.0001 0.7843 -0.0001 -0.0493 0.0000
0.0117 -0.0007 -0.0016 -0.0099 -0.0088
( 33.65%) 0.5801* C 10 s( 20.77%)p 3.81( 79.06%)d 0.01( 0.16%)
0.0003 0.4552 -0.0237 0.0026 -0.3731
-0.0159 -0.8046 -0.0341 0.0506 0.0021
0.0267 -0.0017 -0.0036 -0.0226 -0.0201
4. (1.98452) BD ( 1) N 1 - C 14
( 66.35%) 0.8145* N 1 s( 25.00%)p 3.00( 74.97%)d 0.00( 0.03%)
0.0000 0.5000 -0.0007 0.0000 0.4239
-0.0001 -0.5478 0.0001 -0.5196 0.0001
-0.0096 -0.0091 0.0117 -0.0025 0.0007
( 33.65%) 0.5801* C 14 s( 20.78%)p 3.80( 79.06%)d 0.01( 0.16%)
0.0003 0.4552 -0.0237 0.0026 -0.4350
-0.0185 0.5621 0.0239 0.5330 0.0226
-0.0218 -0.0207 0.0267 -0.0056 0.0016
5. (1.99118) BD ( 1) C 2 - H 3
( 63.47%) 0.7967* C 2 s( 26.42%)p 2.78( 73.53%)d 0.00( 0.05%)
0.0000 0.5140 0.0032 -0.0004 -0.3982
-0.0185 0.5461 -0.0081 0.5271 -0.0170
-0.0097 -0.0102 0.0167 -0.0058 0.0036
( 36.53%) 0.6044* H 3 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 0.0142 -0.0138 -0.0118
In the Second Order Perturbation Theory Analysis table there were no E(2) values over 20 kcal/mol.
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Summary of bond orbitals in molecule below,
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C4H12N)
1. BD ( 1) N 1 - C 2 1.98452 -0.90688 54(v),76(v),134(v),128(v)
130(v),98(v),121(g),123(g)
122(g)
2. BD ( 1) N 1 - C 6 1.98452 -0.90687 99(v),77(v),33(v),133(v)
125(v),131(v),123(g),120(g)
122(g)
3. BD ( 1) N 1 - C 10 1.98452 -0.90684 32(v),98(v),135(v),126(v)
127(v),55(v),123(g),120(g)
121(g)
4. BD ( 1) N 1 - C 14 1.98452 -0.90689 124(v),129(v),132(v),55(v)
33(v),76(v),121(g),120(g)
122(g),77(v),32(v)
Molecular Orbital
[P(CH3)4]+
Optimisation
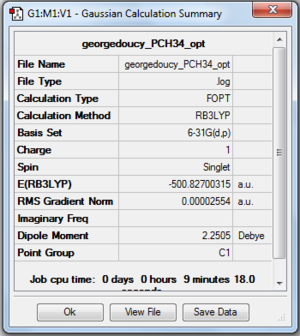
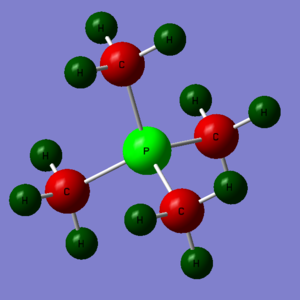
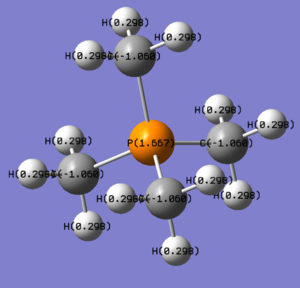
Moving down group 5, the phosphorus ion is analysed (the same basis set and method as the [N(CH3)4]+ ion). P-C bond distance was found to be 1.81664 angstrom and C-P-C bond angle 109.5210. Proof and files below. DOI: 10042\to-http://hdl.handle.net/10042/23868

Item Value Threshold Converged?
Maximum Force 0.000148 0.000450 YES
RMS Force 0.000033 0.000300 YES
Maximum Displacement 0.000894 0.001800 YES
RMS Displacement 0.000305 0.001200 YES
Predicted change in Energy=-1.781935D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
Lowest low frequencies 10% compared to vibrational frequencies and no negative frequencies, optimised molecule. DOI:10042\to-http://hdl.handle.net/10042/23869


Low frequencies --- -0.0023 0.0023 0.0024 52.5732 52.5732 52.5732
Low frequencies --- 188.0647 212.3691 212.3691
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A2 T1 T1
Frequencies -- 188.0647 212.3005 212.3005
Red. masses -- 1.0078 1.0255 1.0255
Frc consts -- 0.0210 0.0272 0.0272
IR Inten -- 0.0000 0.0000 0.0000
Molecular orbital and NBO
Same set up as the [N(CH3)4]+, files below. DOI: 10042\to-http://hdl.handle.net/10042/23891
Summary table for charge distribution of every single atom.
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -1.06015 1.99942 5.05192 0.00882 7.06015
H 2 0.29787 0.00000 0.70109 0.00104 0.70213
H 3 0.29785 0.00000 0.70111 0.00104 0.70215
H 4 0.29784 0.00000 0.70111 0.00104 0.70216
C 5 -1.06016 1.99942 5.05192 0.00882 7.06016
H 6 0.29781 0.00000 0.70114 0.00104 0.70219
H 7 0.29781 0.00000 0.70114 0.00104 0.70219
H 8 0.29786 0.00000 0.70110 0.00104 0.70214
C 9 -1.06006 1.99942 5.05182 0.00883 7.06006
H 10 0.29781 0.00000 0.70115 0.00104 0.70219
H 11 0.29776 0.00000 0.70119 0.00104 0.70224
H 12 0.29781 0.00000 0.70115 0.00104 0.70219
C 13 -1.06016 1.99942 5.05192 0.00882 7.06016
H 14 0.29786 0.00000 0.70110 0.00104 0.70214
H 15 0.29782 0.00000 0.70114 0.00104 0.70218
H 16 0.29781 0.00000 0.70114 0.00104 0.70219
P 17 1.66663 9.99814 3.28858 0.04665 13.33337
=======================================================================
* Total * 1.00000 17.99581 31.90973 0.09446 50.00000
Unlike the nitrogen ion the bond contribution between the P-C bond is different while still showing sp3 character, here roughly 60/40 contribution with the carbon atom (again unlike the nitrogen ion) contributing the most.
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98387) BD ( 1) C 1 - H 2
( 64.79%) 0.8049* C 1 s( 24.88%)p 3.02( 75.07%)d 0.00( 0.04%)
0.0001 -0.4988 0.0070 -0.0005 -0.0799
-0.0217 -0.0050 0.0000 0.8625 0.0032
0.0000 -0.0028 0.0002 -0.0015 -0.0205
( 35.21%) 0.5934* H 2 s( 99.95%)p 0.00( 0.05%)
-0.9998 -0.0008 0.0052 0.0001 -0.0214
2. (1.98384) BD ( 1) C 1 - H 3
( 64.79%) 0.8049* C 1 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0070 0.0005 0.3829
0.0209 0.7041 -0.0019 0.3285 -0.0065
0.0086 0.0038 0.0142 -0.0108 -0.0050
( 35.21%) 0.5934* H 3 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0124 -0.0168 -0.0070
3. (1.98384) BD ( 1) C 1 - H 4
( 64.79%) 0.8049* C 1 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0070 0.0005 0.3799
0.0209 -0.7097 0.0019 0.3200 -0.0065
-0.0086 0.0037 -0.0139 -0.0110 -0.0053
( 35.21%) 0.5934* H 4 s( 99.95%)p 0.00( 0.05%)
0.9998 0.0008 -0.0123 0.0169 -0.0068
4. (1.98030) BD ( 1) C 1 - P 17
( 59.56%) 0.7718* C 1 s( 25.24%)p 2.96( 74.68%)d 0.00( 0.08%)
-0.0002 -0.5021 -0.0171 0.0020 0.8374
-0.0154 -0.0005 0.0000 -0.2128 0.0039
0.0000 0.0120 0.0000 -0.0236 0.0119
( 40.44%) 0.6359* P 17 s( 25.01%)p 2.96( 74.14%)d 0.03( 0.85%)
0.0000 -0.0001 -0.5001 0.0009 0.0000
0.0000 -0.8344 0.0012 0.0000 0.0005
0.0000 0.0000 0.2126 -0.0003 0.0001
0.0382 0.0000 -0.0751 0.0377
5. (1.98386) BD ( 1) C 5 - H 6
( 64.78%) 0.8049* C 5 s( 24.88%)p 3.02( 75.08%)d 0.00( 0.04%)
-0.0001 0.4987 -0.0070 0.0005 0.3779
-0.0110 -0.7107 -0.0165 0.3199 -0.0095
-0.0131 0.0098 -0.0111 -0.0043 -0.0043
( 35.22%) 0.5934* H 6 s( 99.95%)p 0.00( 0.05%)
Again no E(2) values over 20 kcal/mol in the Second Order Perturbation Analysis table.
Second Order Perturbation Theory Analysis of Fock Matrix in NBO Basis
Threshold for printing: 0.50 kcal/mol
E(2) E(j)-E(i) F(i,j)
Donor NBO (i) Acceptor NBO (j) kcal/mol a.u. a.u.
===================================================================================================
Summary of bond orbitals in molecule below.
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C4H12P)
1. BD ( 1) C 1 - H 2 1.98387 -0.69146 135(v)
2. BD ( 1) C 1 - H 3 1.98384 -0.69146 131(v)
3. BD ( 1) C 1 - H 4 1.98384 -0.69146 139(v)
4. BD ( 1) C 1 - P 17 1.98030 -0.78611 131(g),139(g),135(g),129(v)
137(v),133(v)
5. BD ( 1) C 5 - H 6 1.98386 -0.69145 139(v)
6. BD ( 1) C 5 - H 7 1.98384 -0.69143 127(v)
7. BD ( 1) C 5 - H 8 1.98387 -0.69145 135(v)
8. BD ( 1) C 5 - P 17 1.98030 -0.78603 127(g),139(g),135(g),138(v)
132(v),125(v)
9. BD ( 1) C 9 - H 10 1.98389 -0.69141 131(v)
10. BD ( 1) C 9 - H 11 1.98386 -0.69141 127(v)
11. BD ( 1) C 9 - H 12 1.98389 -0.69141 139(v)
12. BD ( 1) C 9 - P 17 1.98029 -0.78585 127(g),131(g),139(g),130(v)
136(v),124(v)
13. BD ( 1) C 13 - H 14 1.98388 -0.69145 135(v)
14. BD ( 1) C 13 - H 15 1.98384 -0.69143 127(v)
15. BD ( 1) C 13 - H 16 1.98386 -0.69145 131(v)
16. BD ( 1) C 13 - P 17 1.98030 -0.78603 127(g),131(g),135(g),128(v)
134(v),126(v)
[S(CH3)3]+
Optimisation
This time the central atom is in group 6 leading to different structure set-up in Gaussview, using a NH3 replacing the hydrogens with methyl groups and finally replacing the nitrogen with sulphur. In the calculation step charge is set to 1 using the same basis set and method as the above ions. Optimised molecule gave a S-C bond distance of 1.82273 angstrom with a C-S-C bond angle of 102.7870.
Proof and files below.
DOI: 10042\to-http://hdl.handle.net/10042/23899
Item Value Threshold Converged?
Maximum Force 0.000128 0.000450 YES
RMS Force 0.000040 0.000300 YES
Maximum Displacement 0.001399 0.001800 YES
RMS Displacement 0.000339 0.001200 YES
Predicted change in Energy=-1.811699D-07
Optimization completed.
-- Stationary point found.
Frequency Analysis
Again to further prove optimisation, lowest low frequencies order of magnitude from the vibrational frequencies. DOI:100042\to-http://hdl.handle.net/10042/23900
Low frequencies --- -19.7927 0.0031 0.0048 0.0051 12.6896 31.1934
Low frequencies --- 162.2679 194.2039 205.0893
Harmonic frequencies (cm**-1), IR intensities (KM/Mole), Raman scattering
activities (A**4/AMU), depolarization ratios for plane and unpolarized
incident light, reduced masses (AMU), force constants (mDyne/A),
and normal coordinates:
1 2 3
A A A
Frequencies -- 162.2251 194.1925 205.0631
Red. masses -- 1.0181 1.0396 1.0387
Frc consts -- 0.0158 0.0231 0.0257
IR Inten -- 0.0005 0.0610 0.0602
Molecular orbital and NBO
DOI:10042\to-http://hdl.handle.net/10042/23901

Summary table for charge distribution of every single atom.
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.84536 1.99943 4.83707 0.00886 6.84536
H 2 0.29733 0.00000 0.70159 0.00107 0.70267
H 3 0.29730 0.00000 0.70162 0.00107 0.70270
H 4 0.27858 0.00000 0.71953 0.00190 0.72142
C 5 -0.84549 1.99943 4.83720 0.00886 6.84549
H 6 0.29729 0.00000 0.70164 0.00107 0.70271
H 7 0.29731 0.00000 0.70161 0.00107 0.70269
H 8 0.27865 0.00000 0.71946 0.00189 0.72135
C 9 -0.84549 1.99943 4.83720 0.00886 6.84549
H 10 0.29732 0.00000 0.70160 0.00107 0.70268
H 11 0.29734 0.00000 0.70158 0.00107 0.70266
H 12 0.27860 0.00000 0.71950 0.00189 0.72140
S 13 0.91661 9.99896 5.03970 0.04474 15.08339
=======================================================================
* Total * 1.00000 15.99724 25.91932 0.08345 42.00000
The sulphur ion is again unlike the other two with a almost 50:50 split of bond contribution between the C-S bond with sulphur holding the majority. The C-S bond shows 20% s character and 80% p character on the sulphur giving sp3d hybridization.
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98720) BD ( 1) C 1 - H 2
( 64.83%) 0.8051* C 1 s( 26.51%)p 2.77( 73.44%)d 0.00( 0.05%)
0.0000 0.5148 -0.0056 0.0006 -0.7753
-0.0063 0.2416 -0.0131 -0.2732 0.0079
-0.0121 0.0130 -0.0075 0.0091 -0.0059
( 35.17%) 0.5931* H 2 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 0.0221 -0.0028 0.0058
2. (1.98721) BD ( 1) C 1 - H 3
( 64.82%) 0.8051* C 1 s( 26.50%)p 2.77( 73.45%)d 0.00( 0.05%)
0.0000 0.5147 -0.0056 0.0006 0.1922
-0.0135 -0.7891 -0.0055 -0.2731 0.0079
-0.0108 -0.0067 0.0135 -0.0105 -0.0059
( 35.18%) 0.5931* H 3 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0011 -0.0014 0.0222 0.0058
3. (1.99412) BD ( 1) C 1 - H 4
( 64.22%) 0.8014* C 1 s( 27.25%)p 2.67( 72.71%)d 0.00( 0.05%)
0.0001 0.5220 -0.0018 0.0010 0.0684
-0.0147 0.0643 -0.0138 0.8472 0.0076
0.0023 0.0065 0.0061 0.0001 0.0193
( 35.78%) 0.5981* H 4 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0035 0.0013 0.0013 -0.0226
4. (1.98631) BD ( 1) C 1 - S 13
( 48.66%) 0.6976* C 1 s( 19.69%)p 4.07( 80.17%)d 0.01( 0.14%)
0.0003 0.4435 0.0140 -0.0033 0.5966
-0.0046 0.5599 -0.0044 -0.3635 -0.0098
0.0265 -0.0175 -0.0164 0.0017 -0.0096
( 51.34%) 0.7165* S 13 s( 16.96%)p 4.86( 82.41%)d 0.04( 0.63%)
0.0000 0.0001 0.4117 -0.0076 0.0012
0.0000 -0.5917 0.0260 0.0000 -0.5557
0.0245 0.0000 0.4039 0.0259 0.0494
-0.0452 -0.0424 0.0031 -0.0051
Again no E(2) value over 20 kcal/mol, leaving the Second Order Perturbation Theory Analysis table.
Summary of bond oritals below.
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C3H9S)
1. BD ( 1) C 1 - H 2 1.98720 -0.72262 109(v),90(v)
2. BD ( 1) C 1 - H 3 1.98721 -0.72258 105(v),90(v)
3. BD ( 1) C 1 - H 4 1.99412 -0.72714
4. BD ( 1) C 1 - S 13 1.98631 -0.83001 102(v),107(v),109(g),105(g)
5. BD ( 1) C 5 - H 6 1.98723 -0.72257 101(v)
6. BD ( 1) C 5 - H 7 1.98722 -0.72253 109(v),89(v)
7. BD ( 1) C 5 - H 8 1.99413 -0.72701
8. BD ( 1) C 5 - S 13 1.98631 -0.82990 106(v),99(v),109(g),101(g)
9. BD ( 1) C 9 - H 10 1.98722 -0.72255 105(v),89(v)
10. BD ( 1) C 9 - H 11 1.98720 -0.72256 101(v)
11. BD ( 1) C 9 - H 12 1.99412 -0.72709
12. BD ( 1) C 9 - S 13 1.98632 -0.83003 103(v),98(v),101(g),105(g)
13. CR ( 1) C 1 1.99942 -10.30644 101(g),32(v),36(v),40(v)
14. CR ( 1) C 5 1.99942 -10.30647 105(g),54(v),58(v),62(v)
15. CR ( 1) C 9 1.99942 -10.30643 109(g),76(v),80(v),84(v)
Comparison
| Ion | C-X Bond Distance/angstrom | C-X-C Bond angle | Bond Contribution C-X(Geometry) |
|---|---|---|---|
| [N(CH3)4]+ | 1.50942 | 109.476 | C:33% N:66%(sp3 tetrahedral) |
| [P(CH3)4]+ | 1.81664 | 109.521 | C:60% P:40%(sp3 tetrahedral) |
| [S(CH3)3]+ | 1.82273 | 102.787 | C:50% S:50% (sp3d trigonal bipyramidal |
From the nitrogen ion to the phosphorous only the central ion has changed, moving down group 5, this correlates with the increase in bond distance and angle as phosphorus is a larger, more diffuse atom. Sulphur being in the same row as phosphorus giving similar bond distances, however a difference in structure having an additional lone pair over phosphorus distorting the bond angle away from 1090.
| Ion | Relative Charge | Charge (Colour Image) |
|---|---|---|
| [N(CH3)4]+ | -0.29507 |  |
| [P(CH3)4]+ | 1.66663 |  |
| [S(CH3)3]+ | 0.91661 |  |
For the charge distribution the [N(CH3)4]+ ion has a negative charge on the central atom the other ions show positive relative charges. With nitrogen being a first row element it holds its electrons very tightly with a high z.eff, phosphorus being the most electropositive element in these three ions with a lower z.eff. In standard [NR4]+ the positive charge is usually depicted on the central nitrogen atom but from the above NBO analysis it can be seen to be spread over the methyl groups, especially on the hydrogens, so while the depiction is useful when drawing mechanisms as nitrogen will take negative charge when forming the NR3, but in this computation it should not be depicted that way.
Part two: Influence of Functional Groups
[N(CH3)3(CH2OH)]+
Optimisation
Making the molecule was done by using a methyl group as the center then using the methyl group replacing the hydrogens gives a tert-butyl group and finally one the outer hydrogens replaced with a O-H group. Optimisation using 6-31G(d,p) method and B3LYP basis set, D-space file and proof and optimisation below. DOI: 10042\to-http://hdl.handle.net/10042/23897


Item Value Threshold Converged?
Maximum Force 0.000074 0.000450 YES
RMS Force 0.000015 0.000300 YES
Maximum Displacement 0.001754 0.001800 YES
RMS Displacement 0.000514 0.001200 YES
Predicted change in Energy=-7.285597D-08
Optimization completed.
-- Stationary point found.
MO and NBO Analysis
DOI: 10042\to-http://hdl.handle.net/10042/23902 Summary table for charge distribution of every single atom.


Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.48795 1.99946 4.47337 0.01512 6.48795
H 2 0.26370 0.00000 0.73529 0.00101 0.73630
H 3 0.26615 0.00000 0.73285 0.00100 0.73385
H 4 0.28539 0.00000 0.71319 0.00142 0.71461
C 5 -0.48796 1.99946 4.47337 0.01512 6.48796
H 6 0.26615 0.00000 0.73285 0.00100 0.73385
H 7 0.26369 0.00000 0.73530 0.00101 0.73631
H 8 0.28539 0.00000 0.71319 0.00142 0.71461
C 9 -0.48858 1.99946 4.47500 0.01411 6.48858
H 10 0.26806 0.00000 0.73083 0.00111 0.73194
H 11 0.26805 0.00000 0.73084 0.00111 0.73195
H 12 0.27393 0.00000 0.72508 0.00100 0.72607
C 13 0.09399 1.99938 3.88266 0.02397 5.90601
H 14 0.23380 0.00000 0.76417 0.00203 0.76620
H 15 0.23380 0.00000 0.76417 0.00203 0.76620
O 16 -0.75650 1.99980 6.74443 0.01228 8.75650
H 17 0.53192 0.00000 0.46487 0.00321 0.46808
N 18 -0.31304 1.99949 5.30633 0.00722 7.31304
=======================================================================
* Total * 1.00000 11.99705 37.89778 0.10517 50.00000
Bond contributions between the C-N atoms 33%/66% between the carbon and nitrogen atoms respectively, nitrogen showing 25% s character and 75% p character leaded to sp3.
(Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.99105) BD ( 1) C 1 - H 2
( 63.20%) 0.7950* C 1 s( 26.20%)p 2.81( 73.74%)d 0.00( 0.05%)
0.0000 0.5119 0.0034 -0.0004 0.0034
0.0002 0.5180 -0.0247 -0.6844 -0.0113
0.0001 -0.0003 -0.0202 -0.0091 0.0067
( 36.80%) 0.6066* H 2 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 0.0000 -0.0099 0.0207
2. (1.99030) BD ( 1) C 1 - H 3
( 63.30%) 0.7956* C 1 s( 26.44%)p 2.78( 73.50%)d 0.00( 0.05%)
0.0000 0.5142 0.0027 -0.0004 0.7173
-0.0117 -0.4675 -0.0083 0.0348 -0.0220
-0.0168 0.0043 -0.0028 0.0101 -0.0110
( 36.70%) 0.6058* H 3 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0006 -0.0178 0.0143 0.0028
3. (1.99082) BD ( 1) C 1 - H 4
( 64.35%) 0.8022* C 1 s( 27.10%)p 2.69( 72.85%)d 0.00( 0.05%)
0.0000 -0.5206 -0.0034 0.0003 0.6958
-0.0124 0.4935 0.0042 -0.0127 0.0206
-0.0170 0.0031 0.0025 -0.0081 0.0109
( 35.65%) 0.5971* H 4 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0015 -0.0187 -0.0136 -0.0023
4. (1.98416) BD ( 1) C 1 - N 18
( 33.36%) 0.5776* C 1 s( 20.28%)p 3.92( 79.55%)d 0.01( 0.17%)
-0.0003 -0.4497 0.0237 -0.0023 0.0187
-0.0008 -0.5170 -0.0207 -0.7256 -0.0311
0.0009 0.0008 -0.0334 0.0119 -0.0200
( 66.64%) 0.8163* N 18 s( 25.21%)p 2.97( 74.76%)d 0.00( 0.03%)
0.0000 -0.5021 0.0016 0.0000 -0.0159
-0.0013 0.4985 0.0005 0.7063 -0.0006
0.0004 0.0004 -0.0140 0.0044 -0.0090
15. (1.99652) BD ( 1) C 13 - O 16
( 34.17%) 0.5846* C 13 s( 23.27%)p 3.29( 76.49%)d 0.01( 0.23%)
0.0000 0.4798 -0.0505 -0.0008 -0.6900
-0.0376 -0.5354 -0.0264 0.0007 0.0000
0.0416 -0.0001 0.0000 0.0071 -0.0239
( 65.83%) 0.8114* O 16 s( 30.38%)p 2.29( 69.54%)d 0.00( 0.08%)
0.0000 0.5511 -0.0076 -0.0028 0.6146
0.0059 0.5636 0.0030 -0.0008 0.0000
0.0198 0.0000 0.0000 -0.0086 -0.0180
16. (1.98120) BD ( 1) C 13 - N 18
( 33.62%) 0.5798* C 13 s( 20.85%)p 3.79( 78.97%)d 0.01( 0.18%)
-0.0002 -0.4563 0.0174 0.0000 -0.7203
-0.0163 0.5202 0.0089 -0.0009 0.0000
0.0357 -0.0001 0.0000 -0.0112 0.0201
( 66.38%) 0.8148* N 18 s( 24.37%)p 3.10( 75.59%)d 0.00( 0.04%)
0.0000 -0.4937 -0.0003 -0.0003 0.7283
-0.0004 -0.4749 -0.0002 0.0008 0.0000
0.0152 0.0000 0.0000 -0.0063 0.0094
No E(2) values over 20 kcal/mol, not of interest.
Below, summary of bond orbitals in molecule.
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C4H12NO)
1. BD ( 1) C 1 - H 2 1.99105 -0.70266 141(v)
2. BD ( 1) C 1 - H 3 1.99030 -0.70303 149(v)
3. BD ( 1) C 1 - H 4 1.99082 -0.69886 145(v)
4. BD ( 1) C 1 - N 18 1.98416 -0.89828 143(v),139(v),93(v),71(v)
146(v),49(v),145(g),141(g)
149(g)
5. BD ( 1) C 5 - H 6 1.99030 -0.70303 149(v)
6. BD ( 1) C 5 - H 7 1.99105 -0.70267 137(v)
7. BD ( 1) C 5 - H 8 1.99082 -0.69886 145(v)
8. BD ( 1) C 5 - N 18 1.98417 -0.89830 142(v),134(v),93(v),71(v)
147(v),27(v),145(g),137(g)
149(g)
9. BD ( 1) C 9 - H 10 1.99100 -0.71052 141(v)
10. BD ( 1) C 9 - H 11 1.99100 -0.71052 137(v)
11. BD ( 1) C 9 - H 12 1.99042 -0.70908 149(v)
12. BD ( 1) C 9 - N 18 1.98421 -0.90678 148(v),49(v),27(v),136(v)
140(v),94(v),137(g),141(g)
149(g)
13. BD ( 1) C 13 - H 14 1.98965 -0.72289 137(v),110(v)
14. BD ( 1) C 13 - H 15 1.98964 -0.72285 141(v),110(v)
15. BD ( 1) C 13 - O 16 1.99652 -1.04307 145(v)
16. BD ( 1) C 13 - N 18 1.98120 -0.90136 150(v),70(v),26(v),48(v)
144(v),135(v),138(v),137(g)
141(g),145(g),111(v)
17. BD ( 1) O 16 - H 17 1.98020 -0.89221 149(v),94(v)
18. CR ( 1) C 1 1.99946 -10.28080 44(v),131(v),40(v),36(v)
19. CR ( 1) C 5 1.99946 -10.28080 66(v),131(v),58(v),62(v)
20. CR ( 1) C 9 1.99946 -10.28964 124(v),88(v),80(v),84(v)
21. CR ( 1) C 13 1.99938 -10.35783 132(v),148(g)
22. CR ( 1) O 16 1.99980 -19.15537 92(v),94(v),97(v)
23. CR ( 1) N 18 1.99949 -14.47249 50(v),28(v),72(v),92(v)
24. LP ( 1) O 16 1.96289 -0.77603 149(v),92(v),146(v),147(v)
121(v),97(v)
25. LP ( 2) O 16 1.95477 -0.48243 147(v),146(v),120(v),95(v)
[N(CH3)3(CH2CN)]+
Optimisation
Making the molecule identical to [N(CH3)3(CH2OH)]+ but with CN instead of OH. DOI:10042\to-http://hdl.handle.net/10042/23903


Item Value Threshold Converged?
Maximum Force 0.000005 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000390 0.001800 YES
RMS Displacement 0.000077 0.001200 YES
Predicted change in Energy=-1.341709D-09
Optimization completed.
-- Stationary point found.
MO and NBO Analysis
DOI:10042\to-http://hdl.handle.net/10042/23903



Summary table for charge distribution of every single atom.
Summary of Natural Population Analysis:
Natural Population
Natural -----------------------------------------------
Atom No Charge Core Valence Rydberg Total
-----------------------------------------------------------------------
C 1 -0.48850 1.99946 4.47437 0.01468 6.48850
H 2 0.28208 0.00000 0.71679 0.00113 0.71792
H 3 0.26947 0.00000 0.72952 0.00101 0.73053
H 4 0.27371 0.00000 0.72531 0.00098 0.72629
C 5 -0.48532 1.99946 4.47118 0.01469 6.48532
H 6 0.27074 0.00000 0.72824 0.00101 0.72926
H 7 0.27074 0.00000 0.72825 0.00101 0.72926
H 8 0.27687 0.00000 0.72217 0.00096 0.72313
C 9 -0.48850 1.99946 4.47437 0.01468 6.48850
H 10 0.26947 0.00000 0.72952 0.00101 0.73053
H 11 0.28208 0.00000 0.71679 0.00113 0.71792
H 12 0.27371 0.00000 0.72531 0.00098 0.72629
C 13 -0.35763 1.99915 4.34260 0.01589 6.35763
H 14 0.30886 0.00000 0.68971 0.00143 0.69114
H 15 0.30886 0.00000 0.68971 0.00143 0.69114
C 16 0.20868 1.99940 3.75874 0.03319 5.79132
N 17 -0.18626 1.99966 5.16587 0.02074 7.18626
N 18 -0.28904 1.99950 5.28314 0.00641 7.28904
=======================================================================
* Total * 1.00000 13.99608 39.87159 0.13233 54.00000
Bond contributions of C-N atom again 33%/66% respectively, sp3 nitrogen as before.
Occupancy) Bond orbital/ Coefficients/ Hybrids
---------------------------------------------------------------------------------
1. (1.98996) BD ( 1) C 1 - H 2
( 64.14%) 0.8009* C 1 s( 26.91%)p 2.71( 73.04%)d 0.00( 0.05%)
0.0000 0.5188 0.0041 -0.0004 0.7488
-0.0135 -0.4107 -0.0088 -0.0167 0.0226
-0.0149 -0.0038 0.0022 0.0121 -0.0110
( 35.86%) 0.5988* H 2 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0010 -0.0192 0.0130 -0.0033
2. (1.99102) BD ( 1) C 1 - H 3
( 63.48%) 0.7967* C 1 s( 26.38%)p 2.79( 73.57%)d 0.00( 0.05%)
0.0000 0.5136 0.0034 -0.0004 -0.0587
0.0021 0.5149 -0.0250 0.6829 0.0117
-0.0019 -0.0024 0.0200 -0.0089 0.0065
( 36.52%) 0.6043* H 3 s( 99.95%)p 0.00( 0.05%)
0.9997 0.0005 0.0012 -0.0096 -0.0211
3. (1.98974) BD ( 1) C 1 - H 4
( 63.66%) 0.7979* C 1 s( 26.51%)p 2.77( 73.44%)d 0.00( 0.05%)
0.0000 -0.5149 -0.0023 0.0003 0.6581
-0.0134 0.5474 0.0061 0.0315 -0.0221
-0.0184 -0.0040 -0.0034 -0.0058 0.0110
( 36.34%) 0.6028* H 4 s( 99.95%)p 0.00( 0.05%)
-0.9997 -0.0005 -0.0160 -0.0164 0.0032
4. (1.98448) BD ( 1) C 1 - N 18
( 33.12%) 0.5755* C 1 s( 20.23%)p 3.94( 79.60%)d 0.01( 0.17%)
-0.0003 -0.4491 0.0244 -0.0023 0.0435
0.0036 -0.5140 -0.0215 0.7269 0.0328
0.0018 -0.0029 0.0336 0.0117 -0.0204
( 66.88%) 0.8178* N 18 s( 25.36%)p 2.94( 74.61%)d 0.00( 0.03%)
0.0000 -0.5035 0.0013 0.0001 -0.0510
0.0011 0.4937 0.0009 -0.7070 0.0008
0.0011 -0.0019 0.0140 0.0045 -0.0092
17. (1.99595) BD ( 1) C 16 - N 17
( 42.68%) 0.6533* C 16 s( 47.95%)p 1.09( 52.03%)d 0.00( 0.02%)
-0.0002 0.6909 -0.0450 -0.0044 0.6289
0.0592 -0.3472 -0.0266 0.0000 0.0000
-0.0114 0.0000 0.0000 0.0068 -0.0084
( 57.32%) 0.7571* N 17 s( 45.15%)p 1.21( 54.49%)d 0.01( 0.36%)
0.0000 0.6707 -0.0407 0.0003 -0.6636
-0.0069 0.3232 0.0044 0.0000 0.0000
-0.0412 0.0000 0.0000 0.0315 -0.0303
18. (1.98640) BD ( 2) C 16 - N 17
( 47.13%) 0.6865* C 16 s( 0.00%)p 1.00( 99.95%)d 0.00( 0.05%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 -0.0001 0.0000 0.9996 -0.0159
0.0000 0.0193 -0.0134 0.0000 0.0000
( 52.87%) 0.7271* N 17 s( 0.00%)p 1.00( 99.59%)d 0.00( 0.41%)
0.0000 0.0000 0.0000 0.0000 0.0000
0.0000 -0.0001 0.0000 0.9979 0.0143
0.0000 -0.0574 0.0278 0.0000 0.0000
Again no E(2) values over 20 kcal/mol, not of interest.
Below, summary of bond orbitals in molecule
Natural Bond Orbitals (Summary):
Principal Delocalizations
NBO Occupancy Energy (geminal,vicinal,remote)
====================================================================================
Molecular unit 1 (C5H11N2)
1. BD ( 1) C 1 - H 2 1.98996 -0.72019 149(v)
2. BD ( 1) C 1 - H 3 1.99102 -0.72199 153(v)
3. BD ( 1) C 1 - H 4 1.98974 -0.72133 157(v)
4. BD ( 1) C 1 - N 18 1.98448 -0.92016 72(v),94(v),147(v),150(v)
155(v),51(v),149(g),153(g)
157(g)
5. BD ( 1) C 5 - H 6 1.99103 -0.72678 153(v)
6. BD ( 1) C 5 - H 7 1.99103 -0.72678 145(v)
7. BD ( 1) C 5 - H 8 1.98974 -0.72524 157(v)
8. BD ( 1) C 5 - N 18 1.98350 -0.92284 156(v),95(v),142(v),151(v)
29(v),73(v),157(g),145(g)
153(g)
9. BD ( 1) C 9 - H 10 1.99102 -0.72199 145(v)
10. BD ( 1) C 9 - H 11 1.98996 -0.72019 149(v)
11. BD ( 1) C 9 - H 12 1.98974 -0.72133 157(v)
12. BD ( 1) C 9 - N 18 1.98448 -0.92016 28(v),94(v),146(v),143(v)
154(v),51(v),149(g),145(g)
157(g)
13. BD ( 1) C 13 - H 14 1.97040 -0.74004 159(v),158(v),153(v),113(v)
160(v),156(g)
14. BD ( 1) C 13 - H 15 1.97040 -0.74004 159(v),158(v),145(v),113(v)
160(v),156(g)
15. BD ( 1) C 13 - C 16 1.98886 -0.91559 158(g),122(v),149(v),154(g)
155(g)
16. BD ( 1) C 13 - N 18 1.97746 -0.92402 160(v),158(v),50(v),148(v)
144(v),152(v),29(v),73(v)
114(v),145(g),153(g)
Comparison
Cyano is an electron withdrawing group and the OH is electron donating this leads to differences in the charge distribution between the molecules. The OH containing ion had a larger relative negative charge on the central nitrogen atom with the cyano being more positive in the center, the carbon attached to the functional group was also a lot more positive in the OH containing ion than the cyano.
References
TlBr3 bond distance reference: J. Glaser, G. Johansson, Acta Chemica Scandinavica, 1982, 36, 125.