Rep:Mod:28072000
NH3 Optimisations
NH3 Summary
| Molecule | NH3 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -56.55776873 a.u. |
| RMS Gradient | 0.00000485 a.u. |
| Point Group | C3V |
| Optimised N-H Bond Length | 1.02 Å |
| Optimised H-N-H Bond Angle | 106° |
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000004 | 0.000450 | YES |
| RMS | Force | 0.000004 | 0.000300 | YES |
| Maximum | Displacement | 0.000072 | 0.001800 | YES |
| RMS | Displacement | 0.000035 | 0.001200 | YES |
Predicted change in Energy = -5.986280D-10
Jmol Dynamic Image
Ammonia Molecule |
Log File
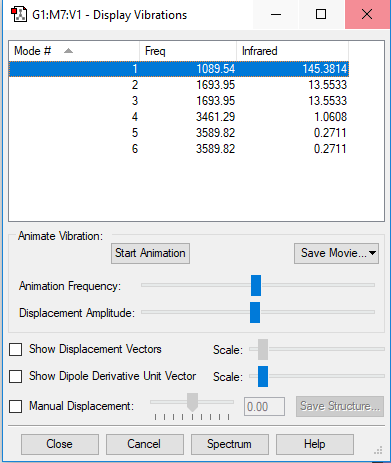
Vibrations and Charges
Display Vibrations
Vibrations, Symmetry & IR Intensity
| Mode# | Wavenumber (cm-1) | Symmetry | IR Intensity | Image |
|---|---|---|---|---|
| 1 | 1090 | A1 | 145 | 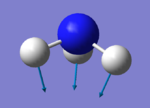
|
| 2 | 1640 | E | 14 | 
|
| 3 | 1694 | E | 14 | 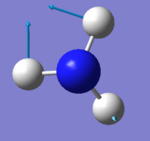
|
| 4 | 3461 | A1 | 1 | 
|
| 5 | 3590 | E | 0 | 
|
| 6 | 3590 | E | 0 | 
|
By the 3N-6 rule since N = 4, you should expect 6 vibrational modes.
Modes 2 and 3 are degenerate; modes 5 and 6 are also degenerate. Modes 1,2 and 3 are bending vibrations. Modes 4,5 and 6 are bond stretch vibrations. Mode 4 is highly symmetrical Mode 1 is the umbrella mode To be IR active there needs to a change in dipole moment. You should expect to see 3 but modes 2 and 3 are degenerate so you only see 2 bands.
Atomic Charges
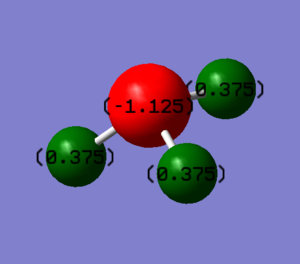
| Atom | Calculated Charge | Oxidation State |
|---|---|---|
| Nitrogen | -1.125 | -3 |
| Hydrogen | +0.375 | +1 |
You would expect Nitrogen to be negatively charged and Hydrogen to be positively charged. This is because the Nitrogen Atom is more electronegative than the Hydrogen Atom. The computational results agree with the expected charges.
Optimised Structures of N2
N2 Summary
| Molecule | N2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -109.52412868 a.u. |
| RMS Gradient | 0.00000060 a.u. |
| Point Group | D∞h |
| Optimised N-N Bond Length | 1.11 Å |
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000001 | 0.000450 | YES |
| RMS | Force | 0.000001 | 0.000300 | YES |
| Maximum | Displacement | 0.000000 | 0.001800 | YES |
| RMS | Displacement | 0.000000 | 0.001200 | YES |
Predicted change in Energy = -3.400991D-13
Jmol Dynamic Image
Nitrogen Molecule |
Log File for Nitrogen
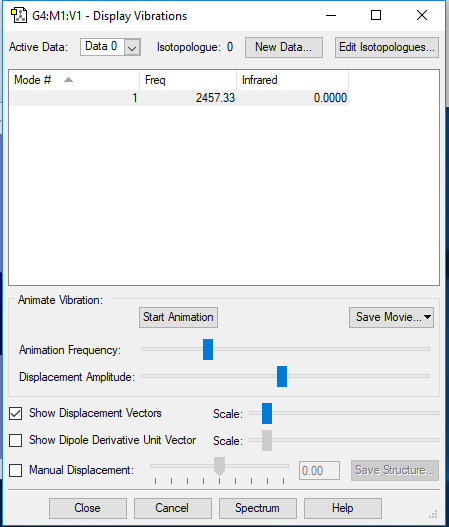
Vibrations and Charges of Nitrogen
In the image above, the frequency corresponds with the wavenumber in cm-1.
The molecule is not IR active because it is a homonuclear diatomic so dipoles cancel out, additionally all vibrations for the molecule is symmetrical so not IR active.
There is no overall charge because the molecule is symmetrical and is a homonuclear diatomic so dipoles cancel out; no overall dipole moment.
Optimised Structures of H2
H2 Summary
| Molecule | H2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -1.17853936 a.u. |
| RMS Gradient | 0.00000017 a.u. |
| Point Group | D∞h |
| Optimised H-H Bond Length | 0.74 Å |
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0 | 0.000450 | YES |
| RMS | Force | 0 | 0.000300 | YES |
| Maximum | Displacement | 0 | 0.001800 | YES |
| RMS | Displacement | 0.000001 | 0.001200 | YES |
Predicted change in Energy = -1.167770D-13
Jmol Dynamic Image
Hydrogen Molecule |
Log File for Hydrogen
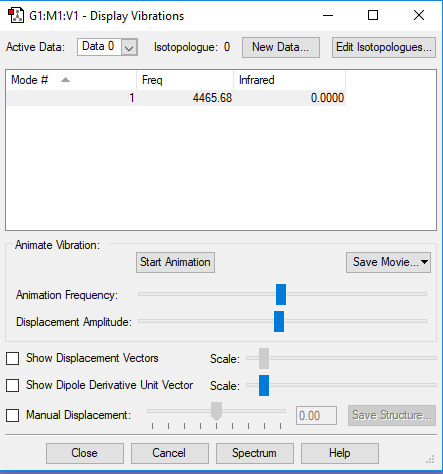
Vibrations and Charges of Hydrogen
The molecule is not IR active because it is a homonuclear diatomic so dipoles cancel out, additionally all vibrations for the molecule is symmetrical so not IR active.
There is no overall charge because the molecule is symmetrical and is a homonuclear diatomic so dipoles cancel out; no overall dipole moment.
Mono-metallic TM complex that coordinates N2
| Unique Identifier | VEJSOV |
| Link to Structure | [Conquest Link ] |
| N≡N Bond Length | 1.116 Å |
| Publication DOI | 10.1002/ejic.201700569 |
| Deposiiton | CCDC 1547291 |
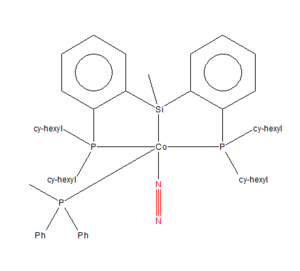
The experimental bond length is 1.12 Å to 3.s.f whereas the computational is 1.11 Å to 3 s.f.. The small difference in the bond lengths is because the N2 which is coordinated to the metal complex, this bond length is impacted by the other ligands bonded to the metal cation and by the metal cation itself. Bonding to the metal cation via a coordinate bond leads to an elongation of the N2 bond. This is because the transiton metal pulls some of the electron density from the N-N bond towards the N-TM bond therefore, weakening and elongating the N-N bond.
Additionally, the N2 in the metal coordinate is not in the gas phase like the other N2 molecule thus impacting the bond length of the two. However, the difference in bond length is negligible (difference is by 0.01 Å). The computational method used is not that accuarate so there are errors that need to be taken into acccount; a better method needs to be used.
Haber-Bosch Process
The Haber-Bosch process is an industrial process which makes Ammonia from the reactants Hydrogen and Nitrogen. The Ammonia can be used for many other processes i.e. for the use of fertilisers.
| E(NH3) | -56.55776873 a.u. |
| 2 x E(NH3) | -113.11553746 a.u. |
| E(N2) | -109.52412868 a.u. |
| E(H2) | -1.17853936 a.u. |
| 3 x E(H2) | -3.53561808 a.u. |
| ΔE = 2 x(NH3) - [E(N2) + 3 xE(H2)] | -0.0557907 a.u. , -146.5 kJ/mol (1 d.p.) |
CO Molecule (Small Molecule)
CO Summary
| Molecule | CO |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -113.30945314 a.u. |
| RMS Gradient | 0.00001828 a.u. |
| Point Group | D∞h |
| Optimised C≡O Bond Length | 1.14 Å |
| Dipole Moment | 0.0599 |
| Item | Value | Threshold | Converged? | |
|---|---|---|---|---|
| Maximum | Force | 0.000032 | 0.000450 | YES |
| RMS | Force | 0.000032 | 0.000300 | YES |
| Maximum | Displacement | 0.000012 | 0.001800 | YES |
| RMS | Displacement | 0.000018 | 0.001200 | YES |
Predicted change in Energy = -3.956716D-10
Jmol Dynamic Image
Carbon Monoxide |
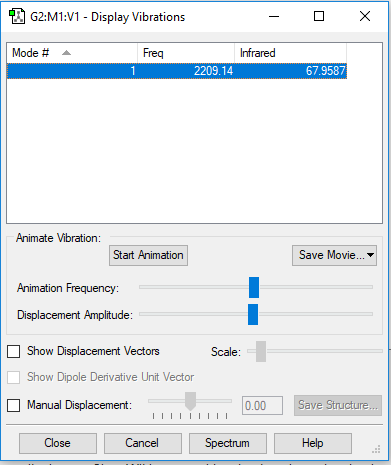
Log File for Carbon Monoxide
Vibrations

Atomic Charges
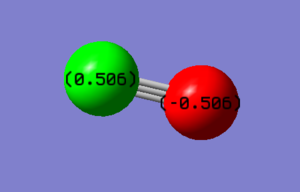
The molecule is linear, but since the Oxygen atom is more electronegative than the Carbon atom that is the reason why the Oxygen atom has a negative atomic charge whereas the Carbon atom has a positive atomic charge.
Mono-metallic TM complex that coordinates CO (Independent Mark)
| Unique Identifier | HEGMAK |
| Link to Structure | [Conquest Link ] |
| C≡O Bond Length | 1.156 Å |
| Publication DOI | 10.1039/C7DT03096G |
| Deposition | CCDC 1562700 |
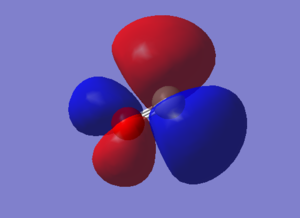
Molecular Orbitals of Carbon Monoxide
The electronic configuration for -
Carbon: 1s2 2s2 2p2 , Oxygen: 1s2 2s2 2p4
In Carbon Monoxide, Carbon is bonded to Oxygen by a triple bond - this means the bonding is made up of 1 σ bond and 2 π bonds. This means that the the 2s orbitals and 2p orbitals are involved in the bonding for this molecule.

Bonding is when there is an interaction between atomic orbitals thus producing a bond whereas the antibonding orbital is the complete opposite as the antibonding orbital doesn't contribute to the bond. Antibonding orbitals contributing to the bond is an unstable and high energy interaction.
HOMO & LUMO Orbitals
HOMO means highest occupied molecular orbital and LUMO means lowest unoccupied molecular orbital. The energies of the HOMO/LUMO are low and are similar to the energies of the bonding 2p and 2s orbitals, they are not high in energy.

- In the image it says a node is change in electron density, that it is a mistake. A node is a region with zero electron density.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES - There is a typo regarding the wavenumber of the 2nd vibration which should be 1694 and not 1640cm-1.
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4/5
Have you completed the calculation and included all relevant information?
YES - You could have commented the vibrational mode rather than just stating it.
Have you added information about MOs and charges on atoms?
YES - You commented on the MO energies in general but missed to explain the relative order of the MOs (e.g. looking at the number of nodes). Only the 1s of O is contributing to the first displayed MO. Therefore it is a non-bonding MO and not a bonding one. The HOMO mainly originating from a in-phase overlap of 2pz orbitals and it is a bonding MO. Otherwise two pairs of areas with the same phase would be observed. The shape of the areas is different from what is expected because s orbitals are mixing in.
The comments on the other MOs are correct but for the 2p orbitals you could have been more specific labelling them (2px,2py and 2pz). For bonding and anti-bonding MOs there are interactions between contributing AOs. For bonding MOs these are constructive and the AOs need to be in phase. For anti-bonding MOs the interactions are destructive and the contributing AOs are out-of-phase. Only for non-bonding MOs there are no interactions between the contributing AOs.
Independence 0.5/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or
YES - You could have explained differences between experimental and computational bond lengths rather than just stating minimal information.
Do an extra calculation on another small molecule, or Do some deeper analysis on your results so far