Rep:Mod:1CSam11
==Computational Lab Experiment 1C==
Part 1
Thermodynamic Analysis of Cyclopentadiene Dimerisation and Hydrogenation Products
- Molecule 1
molecule 1 minimised using MMFF94s force field
| Type of Energy Contribution | Energy (kcal/mol) | Energy (kJ/mol) |
|---|---|---|
| Total Electrostatic Energy | 13.01365 | |
| Total Van der Waal's Energy | 3.54286 | |
| Total Out-of-Plane Bending Energy | 30.77264 | |
| Total Torsional Energy | 2.04136 | |
| Total Stretch Bending Energy | 2.04136 | |
| Total Angle Bending Energy | 0.01486 | |
| Total Bond Stretching Energy | 12.80121 | |
| Total Energy | 55.37344 | 231.838 |
Molecule 2
molecule 2 minimised using MMFF94s force field
| Type of Energy Contribution | Energy (kcal/mol) | Energy (kJ/mol) |
| Total Electrostatic Energy | 58.1907 | 243.633 |
| Total Van der Waal's Energy | 3.46742 | |
| Total Out-of-Plane Bending Energy | 33.19047 | |
| Total Torsional Energy | -2.08217 | |
| Total Stretch Bending Energy | -2.94986 | |
| Total Angle Bending Energy | 0.02197 | |
| Total Bond Stretching Energy | 12.35798 | |
| Total Energy | 14.18488 | |
Molecules 1 and 2 have no partial charges. Of the reactant molecules, number 2, the endo Diels-Alder product, is the most unstable. The Van der Waal’s energy of the endo product is lower than number 1, the exo product, due to two favourable interactions available in the endo product, allowed by virtue of the reduced distance of the overhanging methylene bridging proton two protons of the cyclo-pentene ring. This is illustrated in the figure below:
- Molecule 1 (Exo Product)

| Van der Waal's Interaction | Interaction Length (Å) |
| H1-H5 | 2.632 |
| H2-H5 | 3.448 |
| H3-H5 | 2.667 |
| H4-H5 | 2.668 |
| H3-H6 | 2.622 |
| H4-H6 | 2.624 |
- Molecule 2 (Endo-product)
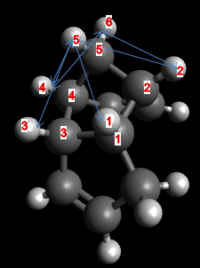
| Van der Waal's Interaction | Interaction Length (Å) |
| H1-H5 | 2.541 |
| H2-H5 | 2.667 |
| H3-H5 | 2.574 |
| H4-H5 | 2.667 |
| H4-H6 | 2.624 |
| H2-H6 | 2.623 |
Torsional and stretch bending energies of the endo product are also lower than for the exo product. Torsional strain is reduced in the endo product due to the gauche-antiperiplanar arrangement of alkyl substituents on bonds C1-C2 and C3-C4, which allows for increased σ-hyperconjugation and reduced steric clashing c.f the gauche-gauche conformation in the exo product. All other energy sources of the endo product are higher. Thus the dimerization is kinetically controlled, as molecule 2, the endo product is favoured over the lower total energy, more stable exo product. This agrees with literature computational modelling reports[2] and experimental observations.[3]
- Molecule 3
molecule 3 minimised using MMFF94s force field
| Type of Energy Contribution | Energy (kcal/mol) | Energy (kJ/mol) |
| Total Electrostatic Energy | 50.72283 | 212.366 |
| Total Van der Waal's Energy | 3.3081 | |
| Total Out-of-Plane Bending Energy | 30.85901 | |
| Total Torsional Energy | -1.92668 | |
| Total Stretch Bending Energy | 0.06933 | |
| Total Angle Bending Energy | 0.01531 | |
| Total Bond Stretching Energy | 13.27678 | |
| Total Energy | 5.12098 | |
Important electrostatic interaction lengths between the nucleophilic allylic carbons and electrophilic overhanging hydrogens are noted below, alongside important H-H Van der Waal's interaction lengths. Numbering of interaction lengths is made with respect to the figure below.
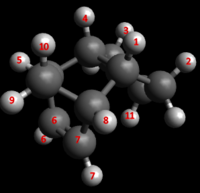
| C-H Electrostatic Interaction | Interaction Lengths (Å) |
| C6-H9 | 2.589 |
| C7-H9 | 2.589 |
| H6-H11 | 2.803 |
| H7-H11 | 2.803 |
| H-H Van der Waal's Interaction | Interaction Lengths (Å) |
| H1-H2 | 2.29 |
| H3-H4 | 2.29 |
| H1-H8 | 2.502 |
| H4-H5 | 2.502 |
| H4-H10 | 2.666 |
| H1-H10 | 2.542 |
| H7-H9 | 3.423 |
| H6-H9 | 3.423 |
- Molecule 4
molecule 4 minimised using MMFF94s force field
| Type of Energy Contribution | Energy (kcal/mol) | Energy (kJ/mol) |
| Total Electrostatic Energy | 41.25749 | 172.737 |
| Total Van der Waal's Energy | 2.82311 | |
| Total Out-of-Plane Bending Energy | 24.68536 | |
| Total Torsional Energy | -1.65719 | |
| Total Stretch Bending Energy | -0.37839 | |
| Total Angle Bending Energy | 0.00028 | |
| Total Bond Stretching Energy | 10.6373 | |
| Total Energy | 5.14702 | |
Important electrostatic interaction lengths between the nucleophilic allylic carbons and electrophilic overhanging hydrogens are noted below, alongside important H-H Van der Waal's interaction lengths. Numbering of interaction lengths is made with respect to the figure below.
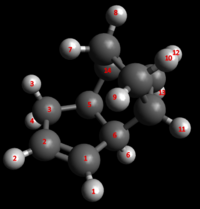
| Electrostatic Interaction | Interaction Distance (Å) |
| C1-H4 | 2.666 |
| C2-H3 | 3.206 |
| C2-H4 | 3.159 |
| C1-H3 | 3.471 |
| H-H Van der Waal's Interaction | Interaction Lengths (Å) |
| H3/4-H5 | 2.285 |
| H1-H6 | 2.674 |
| H5-H14 | 2.459 |
| H5-H13 | 2.518 |
| H6-H13 | 2.632 |
| H8-H12 | 2.609 |
| H10-H12 | 2.572 |
The thermodynamic product of hydrogenation is dimer 4, which has a lower total energy. Van der Waal’s energy for product 4 is lower as only one H-H steric interaction lies within the repulsive region (<2.4 A, [2] and experimental observations.[2]), c.f two steric clashes for product 3. Bond stretch, angle bending, out of plane bending and torsional energies are lower for product 4. Bend stretching energy is lower for product 3. Torsional energy is higher for dimer 3 as an eclipsed arrangement is forced upon the allyl group, with both allylic hydrogens arranged syn-periplanar with the X and Y hydrogens. In molecule 4, the hydrogens of the allylic group are in a staggered conformation with the bonded carbon hydrogens, which reduces steric clashing and increases σ-hyperconjugation, so lowering strain. These effects lower the bond stretch, angle bending and out of plane bending energies as less steric clashing is exhibited. Electrostatic energy of molecule 3 is lower than for molecule 4 as the distance between the partially negative, nucleophilic allylic carbons and the electrophilic overhanging hydrogens is reduced, so allowing for an increased interaction in molecule 3.
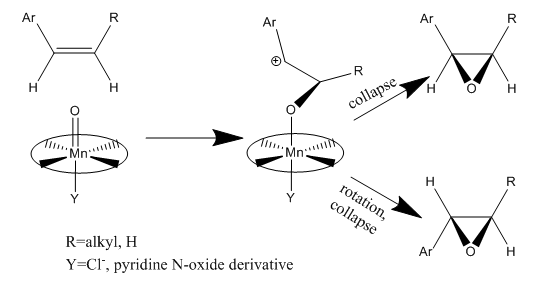
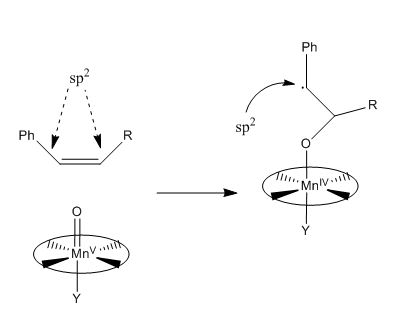
Conformations of an Intermediate in Taxol Synthesis
For both the carbonyl-up and the carbonyl-down atropisomer, four conformational isomers exist, which can be interconverted through a series of bond rotations. These conformational isomers occur due to the stereochemistry of the C ring, which has a cyclohexane structure and four energy minima: chair 1, twist-boat 1, twist-boat 2, chair 2 which are allowed by distortions of the cycloheptane B ring. The cycloheptane B ring has greater strain than cyclohexane C ring, partly due to transannular crowding (steric hindrance by groups on opposite sides of the ring). Thus to find the lowest energy conformation, the cyclohexane ring geometry need only be changed.
Considering only the cyclohexane ring, the planar structure was not trialled as it is well reported that with bond angles of 120º, (10.5º larger than the ideal angle of tetrahedral geometry) angle strain is too high. Additionally, every C-C bond in this structure would be eclipsed, resulting in high eclipsing strains and a highly destabilised structure. Of the reported energetically allowed confromers of the cyclohexane ring: chair; twist-chair; twist-boat and boat structures were trialled.
The highest energy conformer is reported to be the twist-chair form, which could not be modelled for either the atropisomers due to the global energy maximum lying at this geometry, between the two more favourable of the chair (a global minimum) and the twist-boat (local minimum) conformers.
The cyclohexane boat structure has two eclipsed bonds, with severed steric crowding of the "bow" and "stern" hydrogen atoms. As a result of these destabilising effects, which result in the bot form lying at the top of a local energy maxima between the two twist-boat froms, the cyclohexane C ring boat structure could not be found in either the carbonyl-up or carbonyl-down atropisomers.
By twisting the boat conformation, the steric hindrance of the boat is somewhat relieved. This allowed the two twist-boat conformers to be modelled for both atropisomers.
Lifting one carbon atom above the ring plane, and moving the other below ring planereduces the conformer energy further. This staggered structure conformer, in chair conformation, exists with the lowest energy, and is well-reported to exist as the dominant structure adopted by cyclohexane molecules. (REFERENCES: ORGANIC CHEM)
Carbonyl-down Atropisomers
| Conformational Isomer | Conformer 1 | Conformer 2 | Conformer 3 | Conformer 4 | Conformer 5 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | |
| B ring eclipsed, C ring chair 1 |  |
Total Electrostatic Energy = -0.05161
Total Van der Waals = 33.22746 |
||||||||
| B ring staggered, C ring chair 2 | B ring staggered, C ring chair 74.97578 kcal/mol | Total Electrostatic Energy = 0.47721
Total Van der Waals = 35.62172 |
B ring staggered, C ring chair 74.89187 kcal/mol | Total Electrostatic Energy = 0.45760
Total Van der Waals = 35.71917 |
B ring staggered, C ring chair 70.70427 kcal/mol | Total Electrostatic Energy = 0.23104 Total Van der Waals = 35.22782 |
B ring staggered, C ring chair 70.73128 kcal/mol | Total Electrostatic Energy = 0.22888 Total Van der Waals = 35.25229 |
B ring staggered, C ring chair 70.67903 kcal/mol | Total Electrostatic Energy = 0.22795 Total Van der Waals = 35.27567 |
| B ring staggered, C ring twist-boat 1 | B ring staggered, C ring twist boat 138.37778 kcal/mol | Total Electrostatic Energy = 1.91522 Total Van der Waals = 52.42341 |
B ring staggered, C ring twist boat 138.43313 kcal/mol | Total Electrostatic Energy = 1.91704 Total Van der Waals = 52.14369 |
B ring staggered, C ring twist boat 138.43466 kcal/mol | Total Electrostatic Energy = 1.91217 Total Van der Waals = 52.10441 |
B ring staggered, C ring twist boat 65.82924 kcal/mol | Total Electrostatic Energy = 0.09374 Total Van der Waals = 35.38455 |
||
| B ring eclipsed, C ring twist-boat 2 | B ring eclipsed, C ring higher energy twist-boat 75.03719 kcal/mol | Total Electrostatic Energy = 0.40144 Total Van der Waals = 35.79491 |
B ring eclipsed, C ring higher energy twist-boat 68.88626 kcal/mol | Total Electrostatic Energy = -0.04523 Total Van der Waals = 34.66701 |
B ring eclipsed, C ring higher energy twist-boat 66.32321 kcal/mol | Total Electrostatic Energy = -0.04537 Total Van der Waals = 35.08423 |
||||
The conformers, in order of increasing energy and decreasing stability are: chair 1, twist-boat 1, twist-boat 2, chair 2. The lowest energy conformer of the carbonyl-down atropisomer exists as the B ring eclipsed, C ring chair conformation with total energy of 60.55459 kcal/mol. Differences in energy between the optimised structures for each specified conformational isomer is mostly due to the cycloheptane B ring geometry, which can change dramatically with even minute geometry differences of a specified cyclohexane C ring conformer. This is due to increased complexity of the cycloheptane ring, allowing a wide array of geometries lying between two extremes of all-staggered C-C bonds and all-eclipsed C-C bonds. For simplicity here, all structures that did not appear with all-eclipsed bonds were tabulated with B ring staggered bonds.
The lowest energy carbonyl-down confomer exists with a cyclohexane C ring chair geometry, agreeing with literature. (REFERENCE ORGANIC CHEMISTRY) Of the two chair forms, termed chair 1 and chair 2, the lowest energy conformer, chair 1, has an eclipsed B ring structure, c.f. the staggered B ring of the higher energy conformer. This indicates that eclipsing interactions are not the only factor that needs to be considered, as an increase in eclipsing interactions should result in a higher energy conformer. All conformers have a strained gauche arrangement around the cyclohexane ring bridging C-C bond. However, the chair 1 form has a more regular gauche arrangement, as can be seen through dihedral angle analysis. The selected dihedral angles here are those involving the two ring bridging hydrogens, and are as summarised below.
| Conformational Isomer | Torsion Angle H1-C-C-H2 (°) | Torsion Angle H1-C-C-C1 (°) | Torsion Angle H1-C-C-C2 (°) | Torsion Angle H2-C-C-C3 (°) | Torsion Angle H2-C-C-C4 (°) |
|---|---|---|---|---|---|
| B ring eclipsed, C ring chair 1 | 48.5 | 67 | 167 | 68.1 | 163.5 |
| B ring staggered, C ring chair 2 | 52.9 | 62.4 | 168.1 | 59.2 | 174.1 |
| B ring eclipsed, C ring twist-boat 1 | 33.2 | 88.3 | 153.5 | 90.5 | 35 |
| B ring staggered, C ring twist-boat 2 | 28.9 | 86.6 | 147.4 | 86.3 | 142 |
The two chair forms give a more regular gauche arrangement than the twist boat forms, allowing for increasing overlap of σ-σ* orbitals in stabilising hyperconjugation. This effect thus contributes to the increased stabilisation of chair form 1.
Hyperconjugation interactions are summarised in the table below.
| Conformational Isomer | (C-C-(R) σ -> H-C σ*) | (C-H σ -> C-C-(R) σ*) | (C-H σ -> C-H σ*) | (C-C σ -> C-C σ*) | (C-C-(OR) σ -> C-H σ*) | (H-C σ -> C-C-(OR) σ*) | (C-C-(OR) σ -> C-C σ*) | (C-C σ -> C-C-(OR) σ*) |
|---|---|---|---|---|---|---|---|---|
| B ring eclipsed, C ring chair 1 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | O |
| B ring staggered, C ring chair 2 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 1 |
| B ring eclipsed, C ring twist-boat 1 | 0 | 0 | 2 | 2 | 0 | 0 | 1 | 1 |
| B ring staggered, C ring twist-boat 2 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 0 |
As C is more electronegative than H (χP 2.5 c.f 2.1 respectively), so the C–C σ* orbital is lower in energy than the C–H σ* orbital, giving a slightly more favourable hyperconjugative interaction between the C–H σ bond and the C–C σ* orbital compared to that between the C–H σ bond and the C–H σ* orbital (1.5 kcal/mol vs. 1.0 kcal/mol). [1] This effect is increased for the C-C-(R)-> C-C-(OR) σ* interaction, and even more so for the C-C-(OR) σ* -> C-C-(H) σ conjugation due to the increased χP of oxygen relative to carbon and hydrogen. The chair 1 form benefits from both additional stabilisation from the C-(H)σ -> C-C-(OR) σ* and C-H σ -> C-C σ* hyperconjugations, which contribute to the stabilisation of this conformer. The chair 2 conformer benefits from C-H σ -> C-C-(R) σ* and C-C σ -> C-C-(OR) σ* interactions, whilst the twist-boat 1 conformer benefits from C-C σ -> C-C-(OR) σ* conjugations and twist-boat 2 benefits from H-C σ -> C-C-(OR) σ* and C-H σ -> C-C-(R) σ* hyperconjugations. Considering only hyperconjugation effects, the twist-boat 1 would be expected to be less stable than chair 2 and twist-boat 2, with chair-2 being less stable than twist-boat 2. Put simply, using the hyperconjugation analysis, the conformers in theoretical order of increasing energy are: chair 1, twist-boat 2, chair 2, twist-boat 1.
Thus, as the observed energy order differs from that obtained through analysis of equatorial effects (hyperconjugation interactions) it is evident that significant steric hindrance occurs between the B ring carbonyl group and the downward projecting carbon atom of the chair 2 and twist-boat 2 conformers. A 2.757 A close contact interaction exists between the carbonyl-down oxygen and the downward projecting carbon of the chair 2 cylohexane ring. This steric interaction is not present for the chair 1, B eclipsed ring form, as the cyclohexane chair form present here is the mirror image of that present in the B staggered ring isomer, and so the equivalent cyclohexane carbon atom projects upwards. The favourable reduction in the carbonyl-cyclohexane chair steric interaction outweighs the destabilising eclipse strain present in the B-ring, and this is reinforced through analysis of the chair 2 and twist-boat conformers. Both twist-boat conformers would be expected to give a higher energy than both chair forms, considering cyclohexane effects only. However, as is tabulated above, both twist-boats give a lower, more stabilised energy than the chair 2 isomer. This indicates that the carbonyl-cyclohexane repulsive steric clash is the dominant repulsive interaction in the molecule, and is alleviated in the twist-boat forms due to increases in the oxygen-carbon distance by the rotation of the downward projecting cyclohexane carbon atom. The reduction in steric hindrance is greater than the increase in energy obtained from conformation conversion of the C ring from chair to twist-boat.
Twist-boat 2 is higher in energy than twist-boat 1 as, though the principal carbonyl-carbon steric repulsion of chair 2 form is reduced in both twist-boat isomers, twist-boat 2 results in a downward projecting carbon atom, at a distance of 3.128 A from the carbonyl oxygen. In twist boat 1, this carbon projects upwards, and the interaction is reduced due to the longer 4.232 A distance.
The B ring eclipsed, chair 1 form will be used in the following dithiol experimentation as this conformation is the most stable, and therefore will be observed as the predominant conformation. Additionally, the total taxol geometry of the lowest energy conformer is reinforced through comparison with literature, as this conformation was observed as the lowest energy conformation for a dithiol, methylated taxol derivative.
[4]
ChemBio3D Optimisations
As a comparative aside, the twist-boat chair structures were trialled in ChemBio3D using the MMFF94 force field, a maximum of 500 iterations and an RMS gradient of 0.00000001. Results are as tabulated below.
| Conformational Isomer | Conformer 1 | Conformer 2 | ||
|---|---|---|---|---|
| Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | |
| B ring eclipsed, C ring chair 1 | B ring eclipsed, C ring chair 1 61.0278 kcal/mol | Total Energy = 61.0278 | B ring eclipsed, C ring chair 1 60.5445 kcal/mol | Total Energy = 60.5445 |
| B ring staggered, C ring chair 2 | B ring staggered, C ring chair 2 70.6275 kcal/mol | Total Energy = 70.6275 | ||
| B ring eclipsed, C ring twist-boat 2 | B ring eclisped, C ring twist-boat 2 66.3100 kcal/mol | Total Energy = 66.3100 | ||
As for the minimisations performed in Avogadro, the B ring eclipsed, C ring chair 1 returns the lowest total energy, with a 0.00009 kcal/mol difference between the two programs and MMFF94s and MMFF94 force fields. The fit between the twist-boat 2 forms and the chair 2 forms using MMFF94 and MMF94s is similar, with differences of 0.01321 and 0.05153 kcal/mol respectively. All forms return lower energy using the MMFF94 force field. However, the energy distribution between the conformers is the same, with the chair 2 conformer is returned with higher energy than the twist-boat conformer with the MMFF94s force field, reinforcing the results obtained with the MMFF94s force field using Avogadro. This reinforces that the steric interaction between the carbonyl oxygen and the cyclohexane downward projecting carbon in chair form 2 is more repulsive than eclipsing interactions in the B ring obtained through alleviation of steric hindrance by upward rotation of the carbon atom to chair form, and to a reduced effect in the twist-boat 2 form.
Carbonyl-up Atropisomers
Carbonyl-up atropisomer conformations were trialed in the same method as the carbonyl-down atropisomer. The Taxol carbonyl-up atropisomer was modelled with each of the four cyclohexane ring conformations and the total energy minimised using the MMFF94s force field with conjugate gradients in Avogadro. To decrease the energy of each conformer further, atomic geometries were changed slightly prior to further minimisations using the MMF94s force field. The table below summarises a selection of the conformations trialed for each conformational isomer, with the lowest energy geometry for each isomer tabulated.
| Conformational Isomer | Conformer 1 | Conformer 2 | Conformer 3 | Conformer 4 | Conformer 5 | Conformer 6 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | |||
| B ring staggered, C ring chair 2 | B ring staggered, C ring chair 131.2788231 kcal/mol | Total Electrostatic Energy = 1.62706
Total Van der Waals = 47.53841 |
B ring staggered, C ring chair 131.25834 kcal/mol | Total Electrostatic Energy = 1.59637
Total Van der Waals = 47.29274 |
B ring staggered, C ring chair 131.26806 kcal/mol | Total Electrostatic Energy = 1.61176
Total Van der Waals = 47.44958 |
B ring staggered, C ring chair 2 131.07731 kcal/mol | Total Electrostatic Energy = 1.50782
Total Van der Waals =46.74049 |
||||
| B ring eclipsed, C ring chair 1 | B ring eclipsed, C ring chair 1 123.41854 kcal/mol | Total Electrostatic Energy = 1.63300
Total Van der Waals = 44.46712 |
B ring eclipsed, C ring chair 1 123.38535 kcal/mol | Total Electrostatic Energy = 1.63578
Total Van der Waals = 44.47899 |
B ring eclipsed, C ring chair 1 123.37353 kcal/mol | Total Electrostatic Energy = 1.63629
Total Van der Waals = 44.50357 |
B ring eclipsed, C ring chair 1 123.32868 kcal/mol | Total Electrostatic Energy = 1.63859
Total Van der Waals = 44.54910 |
B ring eclipsed, C ring chair 1 123.32349 kcal/mol | Total Electrostatic Energy = 1.63955
Total Van der Waals = 44.57888 |
B ring eclipsed, C ring chair 1 123.32320 kcal/mol | Total Electrostatic Energy = 1.63764
Total Van der Waals = 44.58146 |
| B ring eclipsed, C ring twist-boat 1 | B ring eclipsed, C ring twist-boat 1 128.48151 kcal/mol | Total Electrostatic Energy = 1.66793
Total Van der Waals = 45.98552 |
B ring eclipsed, C ring twist-boat 1 128.47502 kcal/mol | Total Electrostatic Energy = 1.65517
Total Van der Waals = 46.00742 |
B ring eclipsed, C ring twist-boat 1 128.43682 kcal/mol | Total Electrostatic Energy =1.65472
Total Van der Waals = 46.03398 |
B ring eclipsed, C ring twist-boat 1 128.43442 kcal/mol | Total Electrostatic Energy = 1.65733
Total Van der Waals = 46.03176 |
B ring eclipsed, C ring twist-boat 1 128.40590 kcal/mol | Total Electrostatic Energy = 1.65531
Total Van der Waals = 46.02818 | ||
| B ring staggered, C ring twist boat 2 | B ring staggered, C ring twist-boat 135.38221 kcal/mol | Total Electrostatic Energy = 1.61088
Total Van der Waals = 49.00052 |
B ring staggered, C ring twist-boat 135.36156 kcal/mol | Total Electrostatic Energy = 1.60889
Total Van der Waals = 49.13997 |
B ring staggered, C ring twist-boat 2 131.26806 kcal/mol | Total Electrostatic Energy = 1.61029
Total Van der Waals = 47.53394 |
B ring staggered, C ring twist-boat 2 131.27409 kcal/mol | Total Electrostatic Energy = 1.62127
Total Van der Waals = 47.46346 |
B ring staggered, C ring twist-boat 2 131.27882 kcal/mol | Total Electrostatic Energy = 1.62706
Total Van der Waals = 47.53841 |
B ring staggered, C ring twist-boat 2 131.27441 kcal/mol | Total Electrostatic Energy = 1.62048
Total Van der Waals = 47.42292 |
As indicated above, the conformations, in order of increasing energy are as follows:chair 1, twist-boat 1, chair 2, twist-boat 2. This complements the result of the carbonyl-up atropisomer, as for both configurations, the B ring eclipsed, C ring chair conformational isomer exists with lowest energy. As for the carbonyl-up atropisomer, this occurs through the combination of numerous effects, whihc include hyperconjugation and steric hindrance of the electron dense carbonyl oxygen. In order to rationalise electronic stabilisation by hyperconjugation, the summary table is shown below.
| Conformational Isomer | (C-C-(R) σ -> H-C σ*) | (C-H σ -> C-C-(R) σ*) | (C-H σ -> C-H σ*) | (C-C σ -> C-C σ*) | (C-C-(OR) σ -> C-H σ*) | (H-C σ -> C-C-(OR) σ*) | (C-C-(OR) σ -> C-C σ*) | (C-C σ -> C-C-(OR) σ*) |
|---|---|---|---|---|---|---|---|---|
| B ring eclipsed, C ring chair 1 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 0 |
| B ring staggered, C ring chair 2 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 1 |
| B ring eclipsed, C ring twist-boat 1 | 1 | 1 | 0 | 2 | 1 | 1 | 0 | 0 |
| B ring staggered, C ring twist-boat 2 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 1 |
Considering hyperconjugation effects only, the chair 1 and twist-boat 1 conformers would be expected to lie at lower energy and be mores stable than the chair 2 and twist-boat 2 conformers. This is due to the chair and twist-boat 1 conformers obtaining the highly stabilising H-C σ -> C-C-(OR) σ* interaction, compared to the C-C σ -> C-C-(OR) σ* hyperconjugation present in the chair and twist-boat 2 conformers, which exerts a reduced stabilision due to the higher energy C-C σ orbital c.f. the C-H σ orbital, giving reduced overlap, and so weaker orbital interaction and reduced splitting energy with the low energy C-C-(OR) σ* orbital.
Further disparity between the conformers is partially due to steric interations with the bulky, electron dense carbonyl group of the B ring with the C ring carbons. These carbons are bulkier and more electronically dense than the ring hydrogens, which thus do not exert steric repulsions with the carbonyl electron cloud with as great a magnitude as do the carbon atoms. Steric clashing is reduced in the chair 1 isomer, as the eclipsed B ring geometry allows the cyclohexane ring to lie below the plane of the carbonyl, and so the upward projecting cyclohexane carbon, at 3.075 A, is too far away from the carbonyl oxygen to provide any substantial repulsion, whilst the upward projecting B ring carbon, at 4.556 A also does not provide significant steric hindrance. The twist boat 1 form exerts a roughly equivalent steric repulsion through the upward projecting B ring and C ring atoms lying at 4.551 A and 3.056 A from the carbonyl oxygen respectively. The carbonyl steric repulsions present in chair 2 form are increased compared to those present in the chair 1 and twist-boat 1 forms, due to an increase in the number of upward projecting carbon atoms in the B and C rings at close proximity to the carbonyl oxygen. These interactions are roughly equivalent to those present in twist-boat 2 form. In chair 2 form, the C and B ring upward projecting carbons lie at 4.522 and 3.323 A from the carbonyl oxygen. c.f the shorter 4.419 and 3.421 A distances between oxygen and the C and B ring carbons, present in twist-boat 1.
Thus, as both steric effects and hyperconjugation interactions give a roughly equivalent energy for the chair 1, twist-boat 1 and chair 2, twist-boat 2 pairs, the increased stabilisation of each chair form relative to the twist-boat conformers results from the inherent stability of the chair conformation, by virtue of reduced eclipsing interactions and increased σ-σ* overlap, allowing for stronger hyperconjugation interactions.
Comparison of Carbonyl-Up and Carbonyl-Down Atropisomers
The lower energy conformation is the carbonyl-down atropisomer, which at 60.55459 kcal/mol is almost twice as stable as the lowest energy 123.3232 kcal/mol conformation of the carbonyl up atropisomer. Both structures feature a chair C ring, which is energetically favourable and locked into place by the centrally positioned bridge E-cyclononenone , which features heightened non-bonded trans-annular interactions than for the staggered B ring conformers, as has been reported. [5] The lower energy of the carbonyl-down isomer arises from the vast reduction in carbonyl oxygen cyclohexane carbon steric hindrance, which cannot be achieved in the carbonyl-up configuration. As thermodynamically, the final molecule in the oxy-cope equilibrium (carbonyl-up atropisomer) is higher and more unstable than the reagent and first carbonyl-down atropisomer product, this allows for the establishment of an observable equilibrium by the principle of microscopic reversibility.[5] The carbonyl-down atropisomer is an example of a hyperstable olefin, a set of olefins which are stabilised rather than destabilised because of their bridgehead location and are unreactive. Poly-cyclic rings magnify the effects of hyperstability due to the stability gained by the olefin cage structure and the parent poly-cycloalkane being more strained.[6] A selection of successive energy mimisations, achieved using the MMFF94s force field and conjugate gradient algorithm in Avogadro, of the cage structure in question, a bicyclo [6.2.1]undec-1-ene, is tabulated below, and reveals that Oxy-Cope Rearrangement stabilises the molecule from 85.54366 kcal/mol to 60.53455 kcal/mol at the MMFF94s level of theory. The lowest obtained pre-Oxy-Cope geometry and the stabilisation gained upon Oxy-Cope Rearrangement are in accordance with results documented by Paquette et al.[5]
| Taxol Pre-oxy Cope Rearrangement | |||||
|---|---|---|---|---|---|
| Conformer 1 | Conformer 2 | Conformer 3 | |||
| Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) |
| Taxol pre-Oxy Cope Rearrangement minimised to 94.80472 kcal/mol | Total Electrostatic Energy = 19.29532
Total Van der Waals = 38.64676 |
Taxol pre-Oxy Cope Rearrangement minimised to 85.54372 kcal/mol | Total Electrostatic Energy = 16.34218
Total Van der Waals = 37.35729 |
Taxol pre-Oxy Cope Rearrangement minimised to 85.54366 kcal/mol | Total Electrostatic Energy = 16.34231
Total Van der Waals = 37.35759 |
As is the requirement for hyperstable olefins, the parent alkane of the carbonyl-down atropisomer, at 69.5350 kcal/mol using the MMFF94s force field, is higher in total energy than the carbonyl-down olefin, at 60.53445 kcal/mol using the same level of theory and force field.[6]
| Taxol Hydrogenation Product | |||||
|---|---|---|---|---|---|
| Conformer 1 | Conformer 2 | Conformer 3 | |||
| Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) |
| Taxol Hydrogenation Product minimised to 69.54399 kcal/mol | Total Electrostatic Energy = 0.00000
Total Van der Waals = 31.24484 |
Taxol Hydrogenation Product minimised to 69.54357 kcal/mol | Total Electrostatic Energy = 0.00000
Total Van der Waals = 31.21907 |
Taxol Hydrogenation Product minimised to 69.53530 kcal/mol | Total Electrostatic Energy = 0.00000
Total Van der Waals = 31.28088 |
This reinforces the stability of the olefin and the elongated time required for the oxy-cope rearrangement, the product of which is higher in energy than the parent carbonyl-down olefin, as documented above. [5]
Dithiol, Methylated Derivative of Taxol
The lowest energy conformer for taxol, the carbonyl-down, B=eclipsed C=chair isomer, was used as a "starting block" for the dithiol, methylated taxol derivative, drawn using the appropriate tools in Avogadro and minimised using the MMFF94s force field with conjugate gradients in Avogadro. This gave two conformers based on the slope of the dithiol ethylene group. Results are noted below.
| Conformational Isomer | Conformer 1 | Conformer 2 | Conformer 3 | |||
|---|---|---|---|---|---|---|
| Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | |
| B ring eclipsed, C ring chair, dithiol slope right to left | taxol dithiol derivative, carbonyl down, eclipsed, chair, higher energy dithiol conformer | Total Electrostatic Energy = -6.01552
Total Van der Waals = 49.26889 |
taxol dithiol derivative, carbonyl down, eclipsed, chair, lower energy dithiol conformer 100.49638 kcal/mol | Total Electrostatic Energy = -6.04364
Total Van der Waals = 49.35210 |
Total Electronic Energy = 5.07403
Total Van der Waals = 19.97645 | |
| B ring eclipsed, C ring chair, dithiol slope left to right | Dithiol taxol derivative, B ring eclipsed, C ring chair, dithiol slope left to right 100.45058 kcal/mol | Total Electronic Energy = -6.01409 Total Van der Waals = 49.27260 |
Total Electronic Energy = 5.15300 Total Van der Waals = 19.37922 |
|||
This was optimised further to give a lower energy conformation of this isomer.
taxol dithiol derivative, carbonyl down, eclipsed, chair, higher energy dithiol conformer
| Type of Energy Contribution | Energy (kcal/mol) |
| Total Electrostatic Energy | -6.00757 |
| Total Van der Waal's Energy | 38.80345 |
| Total Out-of-Plane Bending Energy | 0.88053 |
| Total Torsional Energy | 9.67023 |
| Total Stretch Bending Energy | 0.67389 |
| Total Angle Bending Energy | 30.97644 |
| Total Bond Stretching Energy | 14.98488 |
| Total Energy | |
- Second Conformer (lower energy)
The lower energy conformer has a geometry in agreement with literature. [1]
NEED DATA FOR THIS
This structure was optimised to a lower energy conformer.
taxol dithiol derivative, carbonyl down, eclipsed, chair, lower energy dithiol conformer
| Type of Energy Contribution | Energy (kcal/mol) |
| Total Electrostatic Energy | -6.01409 |
| Total Van der Waal's Energy | 38.75358 |
| Total Out-of-Plane Bending Energy | 0.88842 |
| Total Torsional Energy | 9.65288 |
| Total Stretch Bending Energy | 0.63018 |
| Total Angle Bending Energy | 30.9487 |
| Total Bond Stretching Energy | 15.07189 |
| Total Energy | 89.93156 |
To check that the B=envelope, C=chair conformer was a minimum for the dithiol, methylated taxol derivative, a number of other isomers were tested, and documented below.
| Conformational Isomer | Conformer 1 | Conformer 2 | Conformer 3 | |||
|---|---|---|---|---|---|---|
| Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | |
| B ring staggered, C ring chair, dithiol slope right to left | taxol dithiol derivative, carbonyl down, eclipsed, twist-boat, dithiol conformer 102.35566 kcal/mol | Total Electrostatic Energy = -7.63857
Total Van der Waals = 51.72894 | ||||
| B ring eclipsed, C ring boat, dithiol slope left to right | taxol dithiol derivative, B ring eclipsed, C ring boat, dithiol left to right 102.28425 kcal/mol | Total Electrostatic Energy = -6.24643
Total Van der Waals = 50.49824 | ||||
| B ring eclipsed, C ring twist-boat, dithiol slope left to right | B ring eclipsed, C ring twist-boat, dithiol left to right 102.58219 kcal/mol | Total Electrostatic Energy = -8.72989
Total Van der Waals = 52.55376 |
B ring eclipsed, C ring twist-boat, dithiol slope left to right | B ring eclipsed, C ring twist-boat, dithiol left to right 102.31850 kcal/mol | Total Electrostatic Energy = -8.72989
Total Van der Waals = 52.55376 | |
NMR Analysis of dithiol Taxol Derivative
Modelling the Sulphur bonded Carbon correction constant
The carbon shift correction for the ketone functionality was calculated using the provided formula in the script. The carbon shift correction for the sulphur bonded carbon was calculated using the method proposed by Braddock et al.[7]
This involved using dimethylsulphide and methane as model NMR complexes, optimising these structures using the MMFF94s force field with conjugate gradient algorithm in Avodagro to total energies of 3.23095 and 0.02638 kcal/mol respectively, which are shown below.
DMS total energy 3.23095 kcal/mol
Methane total energy 0.02638 kcal/mol
These optimised structures were used as input to optimisation and NMR simulation calculations at the DFT B3LYP 6-31G(d,p) level of theory in Gaussian09. This returned optimised DMS and methane structures as shown below, where DMS numbering is documented.
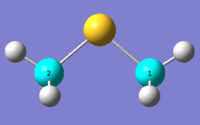
DMS optimised to DFT B3LYP 6-31G(d,p) level of theory Methane optimised to DFT B3LYP 6-31G(d,p) level of theory The DMS and Methane 13C NMR spectra of the optimised DMS and methane structures are as shown.
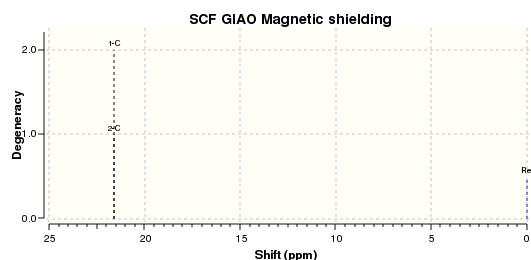
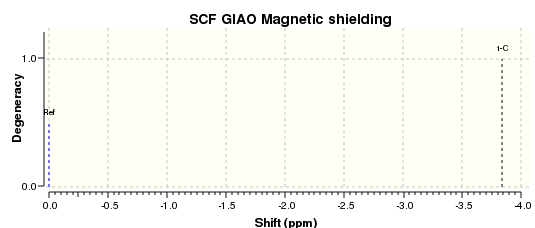
The carbon shift in dimethylsulphide was taken, and from this the methane carbon shift was subtracted, giving the sulphur correction. Parameters involved in this calculation are tabulated below.
| Dimethylsulphide | Methane | δcorr (ppm) | ||
|---|---|---|---|---|
| Carbon Number | δcalc (ppm) | Carbon Number | δcalc (ppm) | |
| C1 | 21.59386267 | C1 | -3.842204108 | 25.43606571 |
| C2 | 21.59386054 | |||
| Average | 21.5938616 | |||
Analysis of Taxol Derivative NMR calculations
An initial NMR at B3LYP 6-31G(d) level of theory was run to compare with that obtained using B3LYP 6-31G(d,p) level of theory. This gave a structure of energy -1651.84055438 a.u, minimised to an RMS Gradient Norm = 0.00000896 a.u. and dipole moment of 1.8244 Debye.
The numbering of this structure used in 13C and 1H NMR spectra analysis is shown below.
Carbon and Hydrogen shifts are as noted.
| 13Carbon NMR | 1H NMR | |||
|---|---|---|---|---|
| Carbon Number | δcalc (ppm) | δcorr (ppm) | Hydrogen Number | δcalc (ppm) |
| C1 | 26.161857 | H20 | 5.3808011259 | |
| C2 | 29.86709953 | H18 | 2.1739491359 | |
| C3 | 147.5631791 | H22 | 1.3884441820 | |
| C4 | 55.88038826 | H23 | 2.0328648211 | |
| C5 | 39.98869248 | H24 | 2.1101413932 | |
| C6 | 212.0205765 | H25 | 1.5686530476 | |
| C7 | 55.208629 | H26 | 1.5772279533 | |
| C8 | 121.0582038 | H27 | 2.2212727976 | |
| C9 | 49.97235076 | H28 | 1.1796658407 | |
| C10 | 24.56279499 | H29 | 0.8114765715 | |
| C11 | 28.49392089 | H30 | 0.5602421860 | |
| C12 | 33.98456806 | H31 | 1.1097325593 | |
| C13 | 66.72226844 | H32 | 0.5843857641 | |
| C14 | 92.90788938 | H33 | 1.0914291282 | |
| C15 | 42.8563047 | H34 | 0.5171291442 | |
| C16 | 25.65950574 | H35 | 2.2921989860 | |
| C17 | 49.53367818 | H36 | 2.3678804139 | |
| C19 | 35.59996883 | H37 | 1.1055670964 | |
| C44 | 46.94962623 | H38 | 0.8801533872 | |
| C45 | 45.28853047 | H39 | 0.9134712328 | |
| H40 | 1.9115711768 | |||
| H41 | 1.9390786429 | |||
| H42 | 1.5109610099 | |||
| H47 | 2.4266532587 | |||
| H49 | 2.3604387856 | |||
| H50 | 2.5928863775 | |||
| H51 | 1.7256286136 | |||
| H52 | 0.8754000000 | |||
| H53 | 0.2364156789 | |||
The hydrogen shifts are more removed from the experimental values than the carbon shifts. L. Paquette, N. A. Pegg, D. Toops, G. D. Maynard, R. D. Rogers, J. Am. Chem. Soc.,, 1990, 112, 277-283. DOI:10.1021/ja00157a043 This is as expected as the two p-orbitals on hydrogen have not been taken into account at this level of theory.
The same molecule was entered into the NMR calculation again, using the expanded DFT B3LYP 6-31G(d,p) force field and basis set. These results, when compared with the 6-31G(d) basis set calculations gave a closer fit to the literature values, L. Paquette, N. A. Pegg, D. Toops, G. D. Maynard, R. D. Rogers, J. Am. Chem. Soc.,, 1990, 112, 277-283. DOI:10.1021/ja00157a043 and those carbons bonded to sulphur and oxygen atoms were entered into sulphur and oxygen corrections. This gave results as follows.
| 13Carbon NMR | ||||||
|---|---|---|---|---|---|---|
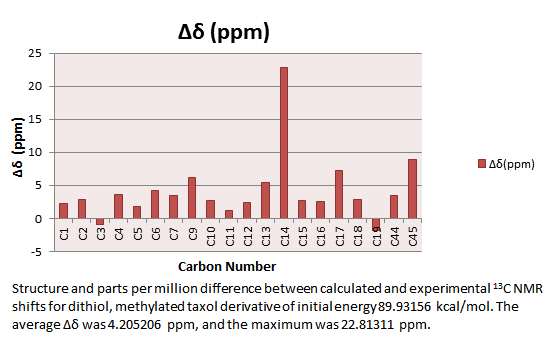
| Carbon Number | δcalc (ppm) | δcorr (0.96δcalc + 12.2) (ppm) | Sulphur Correction (ppm) | δcorr (ppm) | ∆δ(ppm) | Average ∆δ (ppm) |
| C1 | 24.45394017 | 24.45394017 | 2.243940174 | 4.20520568 | ||
| C2 | 28.35963979 | 28.35963979 | 2.799639787 | |||
| C3 | 147.8656084 | 147.8656084 | -0.854391587 | |||
| C4 | 54.93343559 | 54.93343559 | 3.633435588 | |||
| C5 | 38.5101642 | 38.5101642 | 1.730164202 | |||
| C6 | 211.9222905 | 215.6453988 | 215.6453988 | 4.155398844 | ||
| C7 | 54.75635105 | 54.75635105 | 3.45635105 | |||
| C9 | 49.5311262 | 49.5311262 | 6.251126196 | |||
| C10 | 22.58014307 | 22.58014307 | 2.750143067 | |||
| C11 | 26.50329918 | 26.50329918 | 1.153299184 | |||
| C12 | 32.4674164 | 32.4674164 | 2.467416396 | |||
| C13 | 65.93908338 | 65.93908338 | 5.40908338 | |||
| C14 | 92.84098207 | 50.87213142 | 143.7131135 | 22.8131135 | ||
| C15 | 41.47012634 | 41.47012634 | 2.740126336 | |||
| C16 | 24.00755679 | 24.00755679 | 2.617556794 | |||
| C17 | 48.03676504 | 48.03676504 | 7.216765045 | |||
| C18 | 33.69603749 | 33.69603749 | 2.856037493 | |||
| C19 | 33.69603749 | 33.69603749 | -1.773962507 | |||
| C44 | 45.64885345 | 25.43606571 | 71.08491916 | 3.525080837 | ||
| C44 | 44.00772411 | 25.43606571 | 69.44378983 | 8.913789825 | ||
Part 2
Crystallographic Data
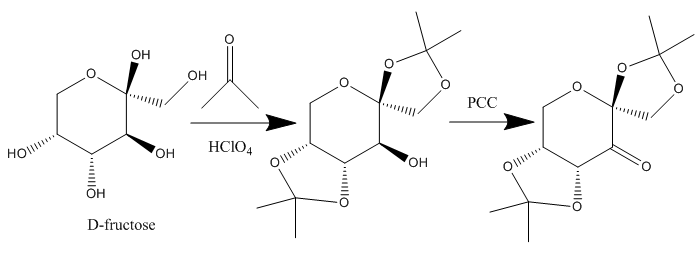
The structure of 21 (Shi asymmetric Fructose Catalyst), synthesised according to the scheme below
was found in the Crystallographic Database using conquest, returning NELQEA
[8] and NELQEA01[9] structures, which were analysed using Mercury. An anomeric centre is a stereocentre resulting from intramolecular acetal (or ketal) formation of an aldehyde (or ketone) group and a hydroxyl group. Two stereochemistries at the anomeric centre are allowed, with each anomer being a diastereoisomers of the other.[10] The α-anomer, with the anomeric substituent axial on the ring is stable and can exist as the dominant isomer over the β-isomer (anomeric substituent equatorial), depending on the other ring substituents. Considering steric arguments only, this is not the expected dominance as in chair conformation, bulky axial substituents cause destabilisation due to unfavourable 1,3-diaxial interactions. The anomeric effect may occur by stabilisation of the axial –OR via donation into the C-O σ* (antibonding) orbital by the lone pair of the pyranose ring oxygen. This hyperconjugation, a stereoelectronic effect acts against, and is observed over steric effects, which render axial -OR stereochemistry as repulsive. The anomeric effect increases in strength with the electronegativity of the anomeric substituent, thus for the OR group, the effect would be expected to be relatively strong in the Shi catalyst. It is reported that the anomeric effect is stronger in pyranose.[11]
For analysis, all C-O bond lengths of the three acetal centres of the Shi catalyst were analysed. The pyranose ring acetal centre displayed bond lengths with a higher equivalence, whilst the acetal centres associated with the 2-dimethyl-(1,3)-dioxolane ring displayed the classical electronic stabilisation of the anomeric centre. This results in one C-O bond being significantly shorter than the other, due to the increase in bond strength of the ring C-O bond associated with π-like overlap of the oxygen p-orbital and the C-O σ*, and the decrease in bonding character of the C-O bond outside the ring, associated with electron donation from the oxygen into the C-O σ*.[11]
The Shi fructose catalyst shows high enantioselectivity for the epoxidation of tri- and trans-substituted olefins. It has been reported that the pyranose oxygen is important for catalytic activity as electronically it inductively activates the carbonyl group to oxone attack due to the high electronegativity of the pyranose oxygen. [9] This effect is reinforced through analysis of the the pyranose ring C-O and 2,2-dimethyl-(1,3)-dioxolane ring C-O bond lengths, which have both axial orientations at the acetal carbon centre, so allowing for the double anomeric effect, where both C-O bonds are stabilised by the anomeric effect. [11]Assuming a double anomeric effect, with equivalent electron donation by each C-O axial lone pair into C-O σ*, one would expect equivalent bond lengths, as observed for molecule 1 in the NELQEA structure. However, NELQEA01 gives a longer 2,2-dimethyl-(1,3)-dioxolane ring C-O σ bond length, which may be indicative of the reported inductive electron withdrawal from the ketone by the pyranose oxygen. This effect increases the electron density of the pyranose C-O bond, increasing the bond strength and reducing bond length. An additional interaction between the ketone C=O π-cloud and 2,2-dimethyl-(1,3)-dioxolane ring C-O σ* bond may also be present, which may reduce electron density at the ketone further through the favourable π-like resonance effect between the C-O σ*-cloud and the C=O π-cloud, which is not present with the pyranose ring C-O bond.
The anomeric O-C-O bond lengths for each structure are noted below.
NELQEA
Shi catalyst crystal structure NELQEA
| Molecule 1 | ' |
| Anomeric Centre bonds | Bond Length (A) |
| C7-01 | 1.413 |
| C7-O2 | 1.441 |
| C10-O5 | 1.409 |
| C10-O4 | 1.439 |
| Molecule 2 | |
| Anomeric Centre Bonds | |
| C19-O7 | 1.38 |
| C19-O8 | 1.443 |
| C22-O11 | 1.416 |
| C22-O10 | 1.438 |
Bond angles of the anomeric centres are as noted below.
| Bond angle | Angle (theta) |
| Molecule 1 | |
| O1-C7-O2 | 103.2 |
| O5-C10-O4 | 105.6 |
| Molecule 2 | |
| O7-C19-O8 | 105.3 |
| O10-C22-O11 | 105.1 |
Non-anomeric bonds and angles were analysed as below.
| Non-anomeric bond lengths | Bond Length (A) |
| Molecule 1 | |
| C2-O6 | 1.403 |
| C2-O2 | 1.403 |
| Molecule 2 | |
| C14-O12 | 1.408 |
| C14-O8 | 1.39 |
| Bond angle | Angle (theta) |
| Molecule 1 | |
| O2-C2-O6 | 113.03 |
| Molecule 2 | |
| O8-C14-O12 | 112.48 |
NELQEA01
This structure was analysed in the same way, with bond lengths and angles as tabulated below.
Shi catalyst crystal structure NELQEA01
| ' | |
| Molecule 1 | |
| Anomeric Centre bonds | Bond Length (A) |
| C10-O5 | 1.428 |
| C10-O4 | 1.456 |
| C9-O1 | 1.423 |
| C9-O2 | 1.454 |
| Molecule 2 | |
| C22-O11 | 1.43 |
| C22-O10 | 1.442 |
| C21-O8 | 1.46 |
| C21-O7 | 1.406 |
| Bond angle | Angle (theta) |
| Molecule 1 | |
| O4-C10-O5 | 105.8 |
| O1-C9-O2 | 103.6 |
| Molecule 2 | |
| O11-C22-O10 | 105.7 |
| O7-C21-O8 | 103 |
Non-anomeric bond lengths and angles are as noted in the following table.
| Non-anomeric bond lengths | Bond Length (A) |
| Molecule 1 | |
| C2-O6 | 1.415 |
| C2-O2 | 1.423 |
| Molecule 2 | |
| C14-O8 | 1.42 |
| C14-O12 | 1.405 |
| Bond angle | Angle (theta) |
| Molecule 1 | |
| O8-C14-O12 | 113.26 |
| Molecule 2 | |
| O6-C2-O2 | 112.78 |
The ketals are at both α-positions, and so act as chiral controls and activating groups. These ketals are sterically bulky enough and in close vicinity to the carbonyl group, allowing for high enantiomeric excess and reaction promotion at the carbonyl group. The lack of any R groups on the 5-membered rings reduces sterics around the carbonyl, allowing for oxone nucleophilic attack. [12]
Molecule 23,[Mn(III)(salen)(Cl)],(1R,2R)-(-)-[Bis(3,5-di-tert-butylsalicylidene)-1,2-cyclohexanediamine]chloromanganese(III), An Intermediate in Jacobsen's Catalyst Synthesis
Two structures of the intermediate ([Mn(III)(salen)(Cl)]) to Jacobsen's catalyst were obtained, TOVNIB01 and TOVNIB02. These are as reported below.
TOVNIB01
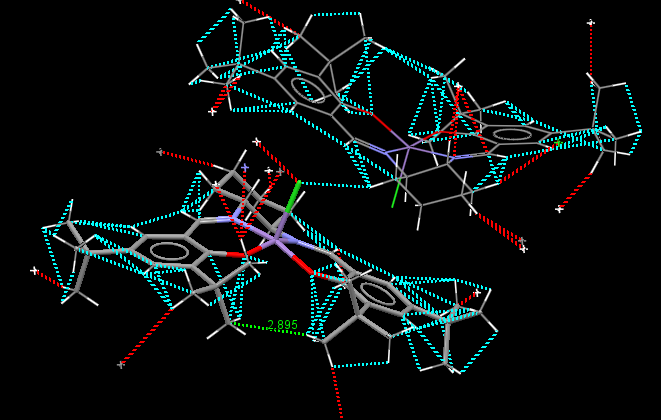
Jacobsen catalyst crystal structure TOVNIB01
TOVNIB02
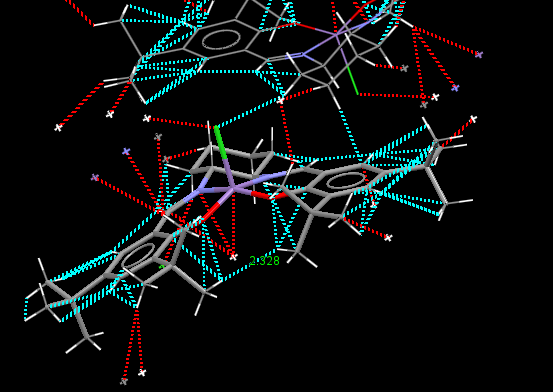
Jacobsen catalyst crystal structure TOVNIB02
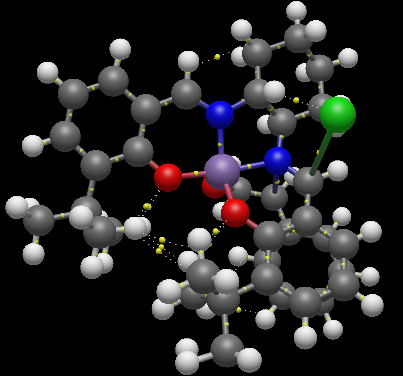
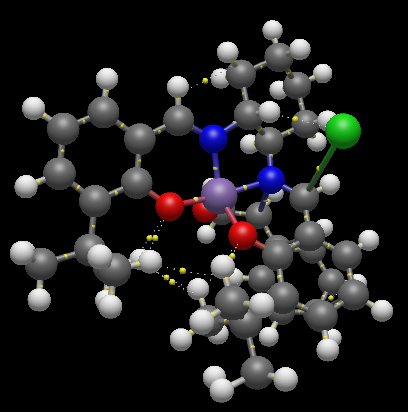
Jacobsen’s ligand, (R,R)- or (S,S)-N,N′-bis(3,5-di-tert-butylsalicylidene)-1,2-cyclohexanediamine, (salen) contains a trans-cyclohexane-1,2-diamine linkage as the sole chiral unit. Salicylidene rings are each bonded to two sterically bulky tert-butyl groups. The enantiopure C2 symmetrical trans-cyclohexane-1,2-diamine serves as a sterodirecting group, creating chiral centres in substrate molecules. Previous X-ray crystal structural studies have shown that metal complexes of the salen ligand exist in an almost planar structure, with the tert-butyl groups of the salicylidene rings not being sufficiently sterically demanding to cause N2O2 coordination distortion. It has been reported that small deviations from planarity may however be required to predetermine the course of asymmetric reactions. [10] As is apparent from the crystal structures TOVNIB01 [13] and TOVNIB02[14] documented, the N2O2 geometry exists with near planarity, indicating that the tert-butyl groups do not have sufficient steric bulk to cause distortion to a stepped geometry, as has been observed for similar Ni(II) complexes.[10] Analysing the close contacts of the tert-butyl groups with the two Mn(III) coordinated oxygens reveals that four interactions are made between each tert-butyl group and each oxygen. Two close contact interactions are made with attack from the chlorine side of the N2O2 coordination plane, with two close contact interactions between each tert-butyl and the oxygen from the under-side of this coordination plane. The tert-butyl groups have sufficient steric bulk to form four close contacts each with one phenyl functionality.
| TOVNIB01 | ' | ' | ' | ' | ' | TOVNIB02 | ' | ' | ' | ' | ' | ||||
| tBu-tBu Interactions | Attack from Cl-side | Attack from under Cl-side | |||||||||||||
| Intramolecular Interactions | Interaction Distance (A) | Intramolecular Interactions | Average Interaction Distance (A) | Intramolecular Interactions | Interaction Distance (A) | Intramolecular Interactions | Average Interaction Distance (A) | Intramolecular Interactions | Average Interaction Distance (A) | ||||||
| TOVNIB01 (C-H, underside of Cl) | 2.895 | tBu(H)-O | 2.193 | tBu(C)-O | 2.884 | tBu(H)-O | 2.55795 | tBu(C)-O | 3.017 | ||||||
| TOVNIB02 (H-H, underside of Cl) | 2.328 | tBu(H)-O | 2.335 | tBu(C)-O | 2.909 | tBu(H)-O | 2.497 | tBu(C)-O | 3.119 | ||||||
Analysis of the Transition State for Shi Catalyst Catalysed Epoxidation of Styrene
To find the styrene transition state in the Shi catalyst catalysed asymmetric epoxidation, the Gibbs free energy of the four transition states given for the R and S-isomers were analysed. This was carried out by accessing the .log file of each transition state structure, where the Gibbs Free Energy was found as the "Sum of Electronic and Thermal Free Energies". Results of the four R and four S-styrene oxide transition states are noted below.
| R-styrene series | ' | ' | S-styrene series | ' | ' |
| Conformer DOI | Gibbs Free Energy (Hartree/Particle) | Gibbs Free Energy +Thermal Correction to Gibbs Free Energy(Hartree/Particle) | Conformer DOI | Gibbs Free Energy (Hartree/Particle) | Gibbs Free Energy + Thermal Correction to Gibbs Free Energy (Hartree/Particle) |
| 10.6084/m9.figshare.822152 | -1303.730703 | -1303.34381 | 10.6084/m9.figshare.823545 | -1303.73383 | -1303.348909 |
| 10.6084/m9.figshare.828520 | -1303.730238 | -1303.344628 | 10.6084/m9.figshare.822136 | -1303.72418 | -1303.338169 |
| 10.6084/m9.figshare.822135 | -1303.736813 | -1303.35075 | 10.6084/m9.figshare.828519 | -1303.72767 | -1303.340216 |
| 10.6084/m9.figshare.822137 | 1303.738044 | 1304.124393 | 10.6084/m9.figshare.826003 | -1303.73850 | -1303.353713 |
This data reveals the structure with the lowest Gibb's Free Energy exists in the R-styrene oxide conformation, which was submitted for NCI and QTAIM analysis.
NCI and QTAIM Analysis of the S-styrene series
Non-covalent interactions refer to intra- and inter-molecular interactions that involve dispersed electromagnetic interactions and do not involve the sharing of electrons. [15] Such interactions are of the order of 1-5 kcal/mol. [15] Non-covalent interactions can be classified into four main categories: Van der Waals forces, π-effects, electrostatic,and hydrophobic effects. For the transition state of the Shi catalyst epoxidation of styrene, the R-styrene oxide conformation provides the lowest Gibb's Free Energy, a result supported by literature observation. [16] [17] [17] [17] Simply, this is due to non-covalent interactions in the transition state, which are greatest for the R-isomer product than for the S-isomer products, leading to the R-isomer pathway having a lower activation energy than the S-isomer pathway, which results in the R-isomer pathway being favoured and so having greater enantiomeric excess experimentally with the Shi catalyst. Both R and S-series transition states operate with a spiro conformation.
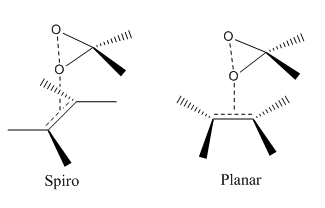
As illustrated below, the spiro A structure is observed for the transition states.
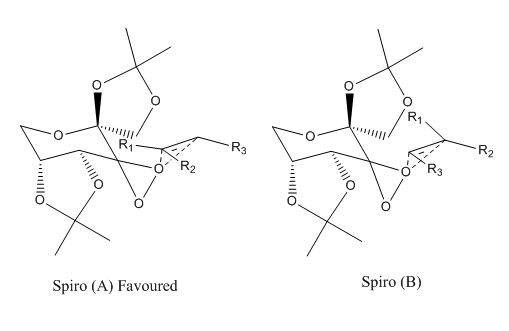
This result complies with literature, and this is favoured over the planar transition state due to the stabilising interaction of the dioxirane oxygen lone pair with the olefin pi* orbital.
[17]
The S-styrene series NCI and QTAIM analysis results are as shown below.
SM11 S isomer of styrene.cub.xyz
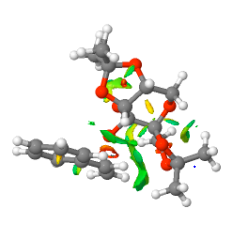
[image:Screenshot NCI 3.PNG|centre|200px|frame|NCI analysis of the transition state for the Shi catalyst catalysed epoxidation towards the S-styrene oxide product 10.6084/m9.figshare.826003]]
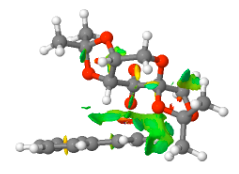
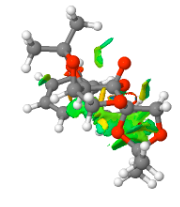
The S-styrene series transition state corresponding to 10.6084/m9.figshare.82600 of Gibbs Free Energy (without thermal correction constant) of -1303.73850 Hartree/Particle was also submitted to QTAIM analysis. Results are as below.
[[image:Qtaim 1.png|centre|200px|frame|Shi catalyst catalysed epoxidation of styrene 10.6084/m9.figshare.826003 S-series]
Shi catalyst catalysed epoxidation of styrene 10.6084/m9.figshare.826003 S-series
QTAIM and NCI analysis revealed the spiro transition state structure. This structure indicates significant C-O bond formation between the oxirane and C1 of the styrene ethyl substituent, which lies at an interaction distance of 1.969 Å, and as revealed by the NCI analysis, can be considered as "half-covalent". This spatial separation is greater than the C-O bond length of the oxirane, which lies at a distance of 1.414 Å and indicates incomplete product C-O bond formation, a conclusion reinforced by the product C-O bond lying at a distance greater than both the epoxide and the hydroxyl C-O typical bond length. The interaction between the β-carbon and the oxirane oxygen occurs over an increased distance of 2.349 Å, and the terminal hydrogen makes an interaction with the ketal oxygen with an interaction distance of 2.489 Å. Additional interactions between the ketal oxygen and the hydrogen at the β-carbon position lies at 2.572 Å. The ketal oxygen makes further interactions with the benzyl ring carbons with strongest interaction distances for each oxygen lying at 3.729 and 5.769 Å. The NCI analysis indicates two major attractive non-covalent interactions. One is intramolecular, and occurs between the eclipsed bonds of the benzene ring and the alkene C=C bond of styrene. The otehr is intermolecular and occurs between the alkene C=C bond of styrene and the oxygens of the dioxirane and the dioxacyclopentane ring which projects closest to the styrene C=C alkene bond.
R-styrene series QTAIM Lowest Energy Transition State
The lowest energy transition state of the Shi catalyst catalysed epoxidation reaction of styrene corresponded to the R-styrene oxide product pathway and was analysed by QTAIM analysis, which revealed the spiro transition state structure, as can be seen in the figures below.



Shi catalyst catalysed epoxidation of styrene lowest energy transition state R-series
This structure indicates significant C-O bond formation between the oxirane and C1 of the styrene ethyl substituent, which lies at an interaction distance of 1.998 Å. This spatial separation is greater than the C-O bond length of the oxirane, which lies at a distance of 1.423 Å and indicates incomplete product C-O bond formation on styrene but significant C-O bond breaking at the dioxirane, a conclusion reinforced by the product C-O bond lying at a distance greater than both the epoxide and the hydroxyl C-O typical bond length. The interaction between the β-carbon and the oxirane oxygen occurs over an increased distance of 2.349 Å, and the terminal hydrogen makes an interaction with the two ketal oxygens with interaction distances of 2.780 and 2.506 Å. Additional interactions between the ketal oxygen and the benzyl ring hydrogen lies at 2.915 and 4.194 Å. The hydrogen of the dioxacyclopentane interacts with the benzyl carbon at an interaction distance of 2.457 Å. The ketal oxygens make further interactions with the benzyl ring carbons with the strongest interaction distances for each oxygen lying at 3.491 and 5.523 Å, and it is these interactions, caused by better overlap of the benzyl ring with the dioxacyclopentane Shi catalyst ring which causes the R-styrene oxide product conformation to be favoured. The S-styrene oxide product pathway causes a significant reduction in the interactions between the benzyl functionality and the ketal oxygens due to the increase in spatial separation, thus causing the higher energies of the transition states for the S-styrene oxide product pathways. In summary, the R-styrene oxide product pathway is favoured as the transition state allows for reduced steric clashing between the bulky phenyl styrene ring and the Shi catalyst dioxacyclopentane ring at the 2-position on the pyranose ring. This also allows for increased interactions between the phenyl ring of styrene and the oxygens of the second dioxacyclopentane functionality at the 4-position on the pyranose ring of the Shi catalyst. Drawing from the NCI analysis of the higher energy S-styrene transition state, the favourable interactions here would be expected to arise between the alkene C=C bond and the oxygens of the dioxirane and dioxacyclopentane, and between the alkene C=C bond and the eclipsed benzene bonds. As the eclipsed benzene-alkene C=C interactions are available to the oxygens of the dioxirane and the dioxacyclopentane, this allows for an increased favourability of interaction for the R-series as the benzene ring is closer to the oxygens of the Shi catalyst, allowing for extended benzene pi-conjugation with the dioxirane and the dioxacyclopentane orbitals of the Shi catalyst. This conclusion is in accord with literature, which reports an attractive interaction between the oxazolidinone moiety of the Shi catalyst and styrene benzyl-π group in the transition state giving the spiro A transition state.[18] Such non-covalent dispersive interactions have been previously reported to influence the stereoselectivity of reactions subject to enantiomeric differentiation.[19]
Analysis of the Transition State for Jacobsen's Catalyst Catalysed Epoxidation of Styrene
The Gibb's Free Energy of each diastereomeric transition state was found from the .log file, with application of the thermal correction to free energy, giving result tabulated below.
| R-styrene series | ' | ' | S-styrene series | ' | ' |
| Conformer DOI | Gibbs Free Energy (Hartree/Particle) | Gibbs Free Energy +Thermal Correction to Gibbs Free Energy(Hartree/Particle) | Conformer DOI | Gibbs Free Energy (Hartree/Particle) | Gibbs Free Energy + Thermal Correction to Gibbs Free Energy (Hartree/Particle) |
| 10.6084/m9.figshare.860441 | -3343.9691970 | -3343.3060230 | 10.6084/m9.figshare.860446 | -3343.960889 | -3343.297060 |
| 10.6084/m9.figshare.860445 | -3343.9631910 | -3343.2987320 | 10.6084/m9.figshare.860449 | -3343.962162 | -3343.297940 |
This analysis revealed that the lowest energy transition state occurred for the R-series, a conclusion supported by literature, [20] [21] [22] and so this transtition state, which corresponds to the cis-breakdown pathway of the radical intermediate shown below, was used in QTAIM analysis.
S-styrene series QTAIM Lowest Energy Transition State
Jacobsen's catalyst catalysed epoxidation of styrene lowest energy transition state S-series
The S-series transition state indicated the significant formation of the C–O bond between the terminal carbon of the ethyl styrene substituent and the (salen)Mn-O oxygen. The C-O bond, at 1.779 Å is greater in length than the Mn-O bond which lies at 1.594 Å. The typical C-O bond length found in hydroxyl bonds lies at 1.43 Å , and 1.47 Å for epoxide C-O bonds. <refname="ja98253389760782">R. C Weast, Handbook of Chemistry and Physics, CRC Press, 65th Edition, 1984 Template:ISBN</ref> As the C-O interaction distance present in the transition is much greater than either of such bond lengths, it can be deduced that C-O bond formation in the transition state is not complete at C1. The β-carbon is oriented by the salen ligand oxygen, with an interaction distance of 3.054 Å. Additionally, the transition state is stabilised by π-overlap between the parallel displaced phenyl functionalities of styrene and the salen ligand, which are oriented with two strong interactions at distances of 3.290 and 3.388 Å.
R-styrene series QTAIM Lowest Energy Transition State
Jacobsen's catalyst catalysed epoxidation of styrene lowest energy transition state R-series
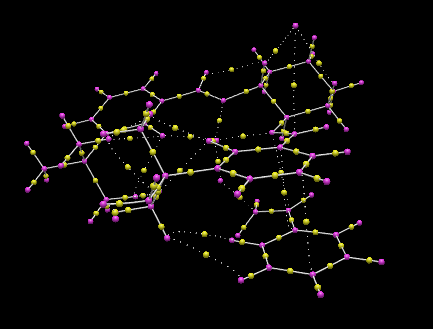
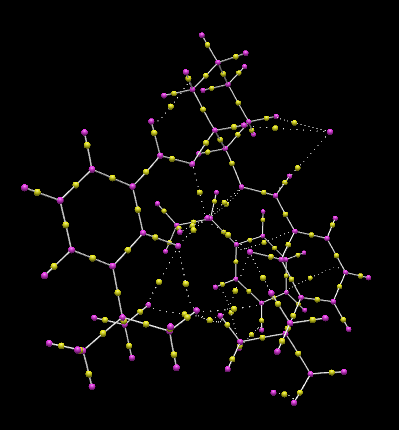
The lowest energy transition state of the Jacobsen’s catalyst catalysed epoxidation reaction of styrene, corresponded to the R-product pathway, and was analysed using QTAIM analysis. This agrees with literature reports carried out with [(R,R)-(–)-N,N-bis(3,5-di-tert-butylsalicylidene)-1,2-cyclohexanediaminomanganese(III) chloride]((R,R)-Jacobsen's catalyst). <refname="ja7696574643">J. Cubillosa, W. Hölderich, Rev. Fac. Ing. Univ. Antioquia, 2007,41, 31-47Template:ISSN</ref> This indicated the significant formation of the C–O bond between the terminal carbon of the ethyl styrene substituent and the (salen)Mn-O oxygen. The C-O bond, at 1.803 Å is greater in length than the Mn-O bond which lies at 1.592 Å. The typical C-O bond length found in hydroxyl bonds lies at 1.43 Å, and 1.47 Å for epoxide C-O bonds. <refname="ja98253389760782">R. C Weast, Handbook of Chemistry and Physics, CRC Press, 65th Edition, 1984 Template:ISBN</ref> As the C-O interaction distance present in the transition is much greater than either of such bond lengths, it can be deduced that C-O bond formation in the transition state is not complete at C1. The β-carbon is oriented by the salen ligand oxygen, with an interaction distance of 3.054 Å. Additionally, the transition state is stabilised by π-overlap between the eclipsed phenyl functionalities of styrene and the salen ligand, which are oriented with two strong interactions at distances of 3.294 and 3.421 Å. The eclipsed benzyl functionalities have an increased spatial separation in the R-styrene oxide product pathway conformation, but have increased overlap of π–clouds as in the S-series transition states, the benzyl rings exhibit parallel displacement, and so reduced stabilisation. Thus the S-styrene oxide product transition states exist at higher energies than the transition states for the R-styrene oxide pathways, disfavouring the S-styrene oxide product at low temperatures and inducing the enantioselectivity for the R-Styrene oxide in the Jacobsen’s catalyst catalysed epoxidation. Additionally, the side-on approach of the Mn=O bond with the styrene oxide is as reported in previous studies, Heterogeneous asymmetric epoxidation of cis-ethyl cinnamte over <refname="ja053389760782">J. A. C. Lobo, Jacobsen’s catalyst immobilized in inorganic porous materials, El Bagre, Kolumbien, 2005</ref> and it can be seen that the non-planarity of the salen-Mn bonding plane reported previously is present in the lowest energy transition state.
Analysis of the Transition State for Shi Catalyst Catalysed Epoxidation of 1,2-Dihydronapthalene
The Gibb's Free Energy of each diastereomeric transition state was found from the .log file, with application of the thermal correction to free energy. This revealed that the lowest energy transition state occurred for the S,R-series, and this was used in QTAIM analysis.
| R,S-series | ' | ' | S,R-series | ' | ' |
| Conformer DOI | Gibbs Free Energy (Hartree/Particle) | Gibbs Free Energy +Thermal Correction to Gibbs Free Energy(Hartree/Particle) | Conformer DOI | Gibbs Free Energy (Hartree/Particle) | Gibbs Free Energy + Thermal Correction to Gibbs Free Energy (Hartree/Particle) |
| DOI:10.6084/m9.figshare.832492 | -1381.120782 | -1,380.697745 | DOI:10.6084/m9.figshare.832538 | -1381.131343 | -1380.709885 |
| DOI:10.6084/m9.figshare.832510 | -1381.125886 | -1,380.704512 | DOI:10.6084/m9.figshare.832536 | -1381.116109 | -1380.696143 |
| DOI:10.6084/m9.figshare.832511 | -1381.134059 | -1,380.711914 | DOI:10.6084/m9.figshare.832545 | -1381.126039 | -1380.704465 |
| DOI:10.6084/m9.figshare.832512 | -1381.126722 | -1,380.703543 | DOI:10.6084/m9.figshare.832544 | -1381.136239 | -1380.714346 |
S,R-Dihydronapthalene oxide series QTAIM Highest Energy Transition State
The transition state of the highest energy S,R-series Shi catalyst epoxidation of 1,2-dihydronapthalene corresponds to the S,R-product pathway. QTAIM analysis allows visualisation of the spiro transition state structure and significant C-O bond formation between the oxirane and C10 of the olefinic bond, which lies at a distance of 2.069 Å. The oxirane oxygen lies at an increased distance of 2.190 Å from C9, which lies closer to the benzyl functionality. QTAIM analysis further reveals the formation of strong interactions between ketal oxygen and the hexane-benzene bridging carbon, which lies at an interaction distance of 3.495 Å, and a much weaker interaction over a distance of 4.826 Å with the C3 ring bridging carbon and the ketal oxygen. Both of these interactions likely involve the overlap between the benzyl π-cloud and oxygen p-orbital lone pairs. A weaker interaction occur between the axial hydrogen of C1 and the pyranose ring oxygen at an interatomic distance of 2.088 Å and a further interaction exists between the C3 ring bridging carbon and the pyranose ring equatorial hydrogen separated by a spatial distance of 2.858 Å. For the transition states corresponding to the R,S-conformation, these interactions are reduced, as the available C1 hydrogen points equatorial, and is thus spatially further from the pyranose oxygen. Additionally, the benzyl group lies beneath the dioxacyclopentane ring at the 2-position on the pyranose. This ring projects down towards the benzyl group, but with the dioxacyclopentane carbon making the closest interaction with the benzyl group of 1,2-dihydronapthalene. This reduces the favourability of this orientation as the carbon lacks p-orbital lone pairs with which it can use to interact with the benzyl π-cloud.
S,R-Dihydronapthalene oxide series QTAIM Lowest Energy Transition State
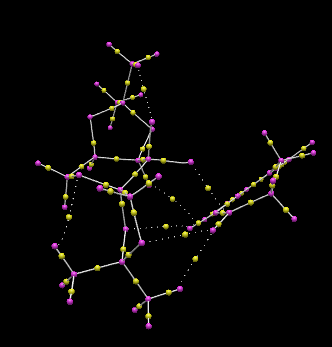
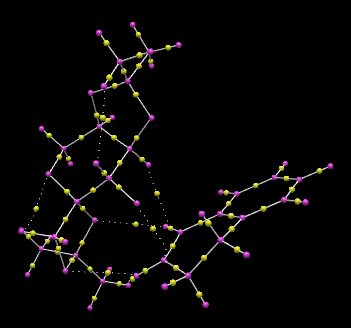

The transition state of lowest Gibb’s free energy of the Shi catalyst epoxidation of 1,2-dihydronapthalene corresponds to the S,R-product pathway, with an endo-structure between the phenyl functionality of the 1,2-dihydronapthalene reagent and the dioxyacyclopentane oxygen of the Shi catalayst. QTAIM analysis allows visualisation of the spiro transition state structure and significant C-O bond formation between the oxirane and C10 of the olefinic bond, which are spatially separated by 2.067 Å. The strong interaction here is due to the upward projection of the C1-C10 bond towards the dioxirane oxygen, increasing availability of the olefinic carbon for attack. The hydrogen of C1 is involved in a hydrogen-bonding like interaction with one oxygen of the dioxyacyclopentane at the C2-position on the pyranose ring, and a Van der Waals radii favourable overlap interaction with the dioxyacyclopentane methyl group. These interactions occur over distances of 2.445 and 2.233 Å respectively. The other dioxyacyclopentane oxygen is involved in an interaction with the other olefinic hydrogen at an interaction distance of 2.496 Å. The dioxirane oxygen lies at an increased distance of 2.189 Å from C9 c.f C10. QTAIM analysis further reveals the formation of strong interactions between ketal oxygen and the hexane-benzene bridging carbon, which lies at an interaction distance of 3.879 Å, and a much weaker interaction over a distance of 5.912 Å with the C3 ring bridging carbon and the other dioxyacyclopentane oxygen. Both of these interactions likely involve the overlap between the benzyl π-cloud and oxygen p-orbital lone pairs. A weaker interaction occur between the axial hydrogen of C2 and the dioxirane oxygen not involved in the epoxide formation, with an interatomic distance of 3.719 Å. For the transition state corresponding to the highest energy S,R-product, with the opposite direction of C10-C1 bond slope, the carbon involved in initial C-O bond formation with the dioxirane points downwards away from the dioxirane oxygen, reducing the interaction with the dioxirane oxygen due to an increased interaction distance of 2.069 Å. Additionally, the formation of the epoxide C-O bond occurs with the other dioxirane oxygen, which reduces steric clashing between the aromatic group of 1,2-dihydronapthalene oxide and the Shi catalyst dioxyacyclopentane rings, and allows for increase electrostatic interactions between the olefin and the dioxyacyclopentane oxygens.
NMR and Modelling of Alkenes and the Associated Epoxides
The two alkenes, and their corresponding epoxides chosen for molecular modelling were styrene (2-phenyloxirane) and 1,2-dihydronapthalene (1,2-dihydronapthalene oxide).
1,2-Dihydronapthalene Oxide
1,2-dihydronapthalene oxide has two chiral centres at each epoxy carbon, as shown in the following figure.

The two chiral centres associated with the epoxide must exist as (R,S) or (S,R) due to the syn-substitution of the epoxy oxygen across the alkene double bond, and thus the two chiral centres can be treated as one chiral centre. One conformational chiral plane arises from the geometry of the C9-C10 bond in the hexane ring, which can either slope up or down with respect to the benzene ring plane. The epoxy chiral centres gives rise to 22 (four) enantiomers, two of which cannot exist favourably due to the syn-oxygen substitution. For each of the two allowed enantiomers, (R,S) and (S,R), two chiral conformations can exist. The chiral conformers can interconvert into each other through chair-antichair inversions of the cyclohexane ring. These are described in the figure below.
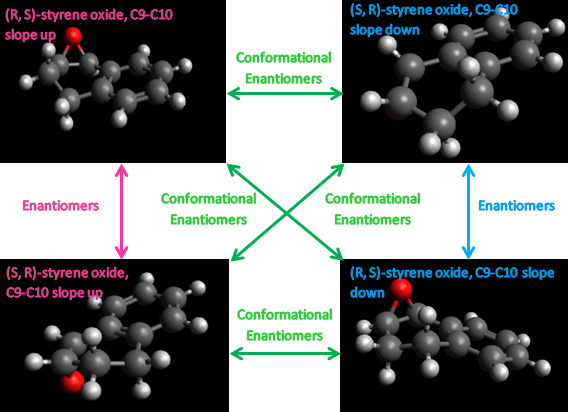
Each of the configurational and conformational isomers was modelled in Avogadro to the MMFF94s, conjugate gradient level of theory. Initial structures were minimised to obtain a minimum energy geometry for each isomer. These are as tabulated below.
| Isomer | Conformer 1 | Conformer 2 | Conformer 3 | |||
|---|---|---|---|---|---|---|
| Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | |
| (S,R)-1,2-dihydronapthalene oxide (C9-C10 slope up) | 1,2-dihydronapthalene oxide 30.68348 kcal/mol | Total Electronic Energy = 5.07558
Total Van der Waals = 19.97117 |
1,2-dihydronapthalene oxide 30.68347 kcal/mol | Total Electronic Energy = 5.07650
Total Van der Waals = 19.95799 |
1,2-dihydronapthalene oxide 30.68364 kcal/mol | Total Electronic Energy = 5.07403
Total Van der Waals = 19.97645 |
| (S,R)-1,2-dihdronapthalene oxide (C9-C10 slope down) | 1,2-dihydronapthalene oxide 30.22445 kcal/mol | Total Electronic Energy = 5.15408 Total Van der Waals = 19.38146 |
1,2-dihydronapthalene oxide 30.22440 kcal/mol | Total Electronic Energy = 5.15300 Total Van der Waals = 19.37922 |
||
| (R,S)-1,2-dihydronapthalene oxide (C9-C10 slope down) | 1,2-dihydronapthalene oxide 30.68345 kcal/mol | Total Electronic Energy = 5.07563 Total Van der Waals = 19.96768 = |
1,2-dihydronapthalene oxide 30.68344 kcal/mol | Total Electronic Energy = 5.07608 Total Van der Waals = 19.96371 |
||
| (S,R)-1,2-dihydronapthalene oxide (C9-C10 slope up) | 1,2-dihydronapthalene oxide 30.22442 kcal/mol | Total Electronic Energy = 5.15329 Total Van der Waals = 19.37677 |
1,2-dihydronapthalene oxide 30.22441 kcal/mol | Total Electronic Energy = 5.15317 Total Van der Waals = 19.37800 |
||
As expected, the conformational isomers, (S,R)-1,2-dihydronapthalene oxide (C9-C10 slope up) and (S,R)-1,2-dihydronapthalene oxide (C9-C10 slope down), and (R,S)-1,2-dihydronapthalene oxide (C9-C10 slope up) and (R,S)-1,2-dihydronapthalene oxide (C9-C10 slope down),give the same energy to within 0.00001 kcal/mol, whilst of the two (S,R) and (R,S)-1,2-dihydronapthalene oxide enantiomers,the (S,R) enantiomer gives a lower energy, and thus is predicted to be more stable than the (R,S) configuration.
NMR
Calculation of Oxide Correction Constants
To allow for the correction factors of the epoxy carbons, ethane and ethylene oxide were modelled using the MMFF94s force field with conjugate gradients in Avogadro. These input structures were optimised to DFT B3LYP level of theory using the 6-31G(d,p) basis set, and NMR calculations carried out on the optimised structures.
- Ethane
The MMFF94s optimised structure used as input to the DFT B3LYP level of theory optimisation is shown below.
Ethane geometry optimised using MMFF94s force field
Ethane |
Energy parameters associated with this structure are as tabulated.
| Energy Contribution | Energy (kcal/mol) |
|---|---|
| Total Electrostatic Energy | 0.00000 |
| Total Van der Waals Energy | 0.11175 |
| Total Out-of-Plane Bending Energy | 0.00000 |
| Total Stretch Bending Energy | -0.00153 |
| Total Torsional Energy | -4.95900 |
| Total Angle Bending Energy | 0.01595 |
| Total Bond Stretching Energy | 0.00742 |
| Total Energy | -4.82541 |
- Ethylene oxide
Ethylene oxide was modelled using the MMFF94s force field in Avogadro, optimised to the DFT B3LYP 6-31G(d,p) level of theory with an NMR simulation carried out. The structure used as DFT B3LYP optimisation input is demonstrated below.
Ethylene oxide geometry optimised using MMFF94s force field
Ethylene oxide |
Energy parameters associated with this structure are tabulated below.
| Energy Contribution | Energy (kcal/mol) |
|---|---|
| Total Electrostatic Energy | 3.49639 |
| Total Van der Waals Energy | 0.01963 |
| Total Out-of-Plane Bending Energy | 0.00000 |
| Total Stretch Bending Energy | -0.00982 |
| Total Torsional Energy | 2.35781 |
| Total Angle Bending Energy | 0.18271 |
| Total Bond Stretching Energy | 0.00462 |
| Total Energy | 6.05133 |
Correction Factors
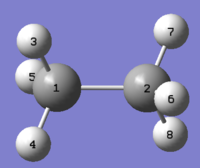
The ethane and ethylene structures gave 13C NMR peaks corrected with reference: TMS B3LYP/6-31G(d,p) Chloroform of shielding 192.17 ppm with NMR Degeneracy Tolerance of 0.05. Numbering used when referring to ethane atom shifts are made with refernce to the figure below.
The calculated C-O oxide δcorr (ppm) was given by subtracting the ethylene shift from the ethane shift for the equivalent atoms and are as documented in the table below.
| Ethane 6-31G(d,p) | Ethylene 6-31G(d,p) | ||||||
|---|---|---|---|---|---|---|---|
| Carbon Number | δcalc (ppm) | Average δcalc (ppm) | Carbon Number | δcalc (ppm) | Average δcalc (ppm) | δcorr (ppm) | |
| C1 | 9.4675 | 9.4675 | C1 | 41.194 | 41.19395 | 31.72645 | |
| C2 | 9.4675 | C2 | 41.1939 | ||||
The equivalent analysis for 1H NMR spectra corrected with reference: TMS B3LYP/6-31G(d,p) Chloroform of shielding reference 31.7462 ppm and NMR Degeneracy Tolerance of 0.05
| Ethane 6-31G(d,p) | Ethylene 6-31G(d,p) | ||||||
|---|---|---|---|---|---|---|---|
| Hydrogen Number | δcalc (ppm) | Average δcalc (ppm) | Hydrogen Number | δcalc (ppm) | Average δcalc (ppm) | δcorr (ppm) | |
| H4 | 0.9506 | 0.949933333 | H1 | 2.6096 | 2.6096 | 1.659666667 | |
| H7 | 0.9504 | H2 | 2.6096 | ||||
| H5 | 0.9504 | H3 | 2.6096 | ||||
| H3 | 0.9488 | H4 | 2.6096 | ||||
| H6 | 0.9488 | ||||||
| H8 | 0.9504 | ||||||
It is well reported that the 1H NMR spectrum of ethane has one signal which is an average of all hydrogen signals in the rapidly interchanging conformations adopted by ethane in solution occurring by rotation around the central C-C bond. [23] However the presently calculated spectrum exhibits three different signals, one for each equivalent hydrogen pair in ethane. This reinforces the static nature of the simulation, which takes into account only the one conformation. In this case, this conformer is the lowest energy staggered conformation, which provides the most accurate representation of the molecule as this conformer, residing at an energy minimum, is the dominant conformation found for a selection of ethane molecules at any given time. The six signals were averaged to give one signal. This provides a rather poor representation of the conformations in solution however, as the experimental final shift is an average of those observed for each conformation. The δcorr (ppm) was given by subtracting the ethylene shift from the ethane shift.
1,2-dihydronapthalene oxide NMR
The NMR simulation was carried out on the lowest energy conformer as this structure represents the dominant structure at any one time in solution. Referring to the table above, this gives the 30.22440 kcal/mol isomer.
Higher Level of Theory
The NMR was also carried out using the DFT B3LYP cc-pVTZ level of theory.This gave a structure with thermal free energies of:
Sum of electronic and thermal Energies= -462.280088
Sum of electronic and thermal Enthalpies= -462.279
Sum of electronic and thermal Free Energies= -462.320756
The 13C NMR and 1H NMR spectra were corrected for TMS B3LYP/cc-pVTZ GIAO are as shown below, respectively.


These results were compared with literature for the (+)-enantiomer, [24] and disparity with experimental shifts calculated and tabulated below.
| Carbon Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| C1 | 26.3666 | 22.2 | 4.1666 | 5.14473 | 8.1021 | 4.1666 | -0.30897 | -23.77355 | |
| C2 | 60.3019 | 55.5 | 31.72645 | 4.8019 | -23.77355 | ||||
| C3 | 57.688 | 55.2 | 31.72645 | 2.488 | -23.47355 | ||||
| C4 | 141.0021 | 132.9 | 8.1021 | 8.1021 | |||||
| C5 | 145.0906 | 137.1 | 7.9906 | 7.9906 | |||||
| C6 | 29.297 | 24.8 | 4.497 | 4.497 | |||||
| C7 | 133.4602 | 128.8 | 4.6602 | 4.6602 | |||||
| C8 | 133.5733 | 128.8 | 4.7733 | 4.7733 | |||||
| C9 | 131.242 | 126.5 | 4.742 | 4.742 | |||||
| C10 | 135.1256 | 129.9 | 5.2256 | 5.2256 | |||||

The carbon NMR spectrum was compared with literature for the (-)-enantiomer, [25] giving results tabulated below.
| Carbon Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| C1 | 26.3666 | 20.8 | 5.5666 | 6.77473 | 9.5021 | 5.5666 | 1.32103 | -22.37355 | |
| C2 | 60.3019 | 54.1 | 31.72645 | 6.2019 | -22.37355 | ||||
| C3 | 57.688 | 51.7 | 31.72645 | 5.988 | -19.97355 | ||||
| C4 | 141.0021 | 131.5 | 9.5021 | 9.5021 | |||||
| C5 | 145.0906 | 135.6 | 9.4906 | 9.4906 | |||||
| C6 | 29.297 | 23.4 | 5.897 | 5.897 | |||||
| C7 | 133.4602 | 127.3 | 6.1602 | 6.1602 | |||||
| C8 | 133.5733 | 127.4 | 6.1733 | 6.1733 | |||||
| C9 | 131.242 | 125.1 | 6.142 | 6.142 | |||||
| C10 | 135.1256 | 128.5 | 6.6256 | 6.6256 | |||||
The calculated carbon shifts have a 1.63 ppm larger disparity with the (-)-enantiomer [25] than with the (+)-enantiomer [24] literature carbon shifts. The disparity between literature shifts and the calculated shifts is due to the static nature of the molecule being run in the calculation, which is of one configuration and conformation. In reality the molecule is dynamic, and making transitions between different energetically available conformers on the submillisecond timescale, and so the different proton shifts that occur in each conformer are completely averaged out on the NMR timescale, which is ~1 sec. Thus a spectrum that is an average of the exchanging states is yielded. The interconversion between conformers is fast on the NMR time scale as it occurs at a much greater rate than the frequency difference between the two conformers. The difference in frequency depends on the Larmor frequency of the observed nucleus and the magnetic field strength, thus changing the magnetic field strength and cooling the solution undergoing the NMR experimental analysis would allow recreation of the static calculated spectrum of one conformer which lies at the energy minimum.
Additionally, intermolecular coupling between molecules is not taken account of in the calculated spectrum, which considers only one molecule. Thus coupling constants and peak multiplicity in the observed spectra would be expected to differ with those present in the calculated spectra.
The equivalent analysis was undertaken for the 1H NMR spectrum. Below is documented the calculated 1H NMR shifts calcualted at the DFT B3LYP cc-pVTZ level of theory and their disparity with literature shifts for the (+)-enantiomer
| Hydrogen Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| H11 | 7.4035 | 7.018 | 0.3855 | 0.281076667 | 0.8442 | 0.3855 | -0.13544 | -2.122333333 | |
| H12 | 7.5534 | 7.1754 | 0.378 | 0.378 | |||||
| H13 | 7.5182 | 7.1356 | 0.3826 | 0.3826 | |||||
| H14 | 7.7166 | 7.322 | 0.3946 | 0.3946 | |||||
| H15 | 2.5507 | 2.2555 | 0.2952 | 0.2952 | |||||
| H16 | 2.8556 | 2.715666667 | 0.139933333 | 0.139933333 | |||||
| H17 | 2.3642 | 1.52 | 0.8442 | 0.8442 | |||||
| H18 | 1.6547 | 1.6988 | -0.0441 | -0.0441 | |||||
| H20 | 3.6844 | 3.667666667 | 1.659666667 | 0.016733333 | -2.008 | ||||
| H21 | 3.8001 | 3.782 | 1.659666667 | 0.0181 | -2.122333333 | ||||
Th 1H NMR shift were compared with literature for the (-)-enantiomer.[25] Results are as tabulated below.
| Hydrogen Number | δcalc (ppm) | Average δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| H11 | 7.4035 | 7.01 | 0.3935 | 0.19252 | 0.0142 | 0.3935 | -0.223996667 | -2.120333333 | ||
| H12 | 7.5534 | 7.5358 | 7.1356 | 0.4178 | 0.4178 | |||||
| H13 | 7.5182 | 7.5358 | 7.1356 | 0.3826 | 0.3826 | |||||
| H14 | 7.7166 | 7.34 | 0.3766 | 0.3766 | ||||||
| H15 | 2.5507 | 2.45 | 0.1007 | 0.1007 | ||||||
| H16 | 2.8556 | 2.675 | 0.1806 | 0.1806 | ||||||
| H17 | 2.3642 | 2.35 | 0.0142 | 0.0142 | ||||||
| H18 | 1.6547 | 1.65 | 0.0047 | 0.0047 | ||||||
| H20 | 3.6844 | 3.65 | 1.659666667 | 0.0344 | -1.990333333 | |||||
| H21 | 3.8001 | 3.78 | 1.659666667 | 0.0201 | -2.120333333 | |||||

A Higher Energy Enantiomer, 30.22441 kcal/mol input energy
To allow for a comparison of the lowest energy isomer NMR, an additional NMR of a higher entropy isomer was carried out on (S,R)-1,2-dihydronapthalene oxide, of input energy 30.22441 kcal/mol, modelled at the DFT B3LYP 6-31G(d,p) level of theory in chloroform solvent, giving the structure shown below.
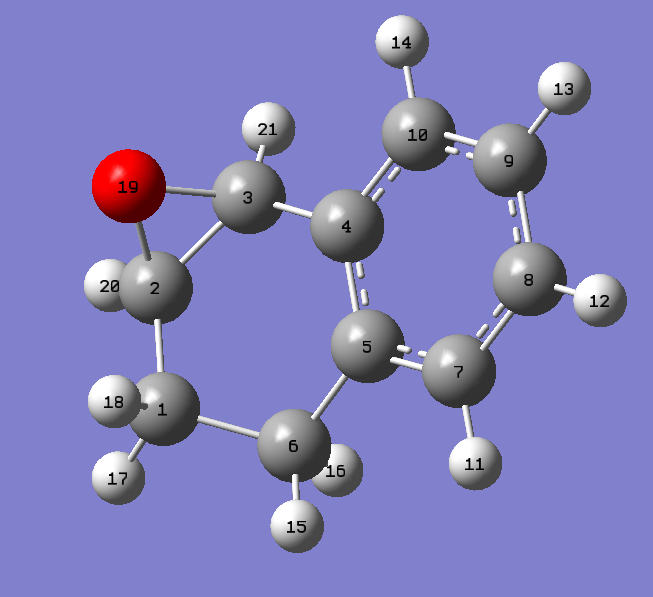

Following TMS/DFT B3LYP 6-31G(d,p) (chloroform) correction application, this gave 1H and 13C NMR spectra as documented below.
1H NMR data
Comparison of B3LYP 6-31G(d,p) optimised structure with MMFF94s force field, conjugate gradient optimisation algorithm
The 30.2441 kcal/mol (S,R)-1,2-dihydronapthalene oxide conformer was optimised through the HPC using Gaussian 09 to the DFT B3LYP 6-31G (d,p) level of theory, and NMR, IR and VCD simulations run on this optimised configuration. Energetics of the optimised molecule are reported in the table below. The lower total energy of the molecule when run to the DFT B3LYP level of theory is lower than at the MMFF94s, conjugate gradient level of theory.
| Energy Contribution | Energy (kcal/mol) |
|---|---|
| Total Electrostatic Energy | -4.01005 |
| Total Van der Waals Energy | 18.10016 |
| Total Out-of-Plane Bending Energy | 0.01243 |
| Total Stretch Bending Energy | -0.46660 |
| Total Torsional Energy | 1.92867 |
| Total Angle Bending Energy | 3.35949 |
| Total Bond Stretching Energy | 3.11745 |
| Total Energy | 22.04156 |
Comparison of 1H NMR data with (+)-1,2-Dihydronapthalene Oxide Literature
Comparing the calculated NMR shifts with D. Xiong et al. (USE REFERENCE IN FIGURE), the disparity between calculated and experimental shifts could be found. This is reported in the following table and figure.
| Hydrogen Number | δcalc (ppm) | δexp (ppm) | ∆δ (ppm) |
|---|---|---|---|
| H11 | 7.251492731 | 7.01 | 0.241492731 |
| H12 | 7.391151507 | 7.18 | 0.211151507 |
| H13 | 7.387571667 | 7.185 | 0.202571667 |
| H14 | 7.615118252 | 7.34 | 0.275118252 |
| H15 | 2.267285883 | 2.45 | -0.182714117 |
| H16 | 2.946631263 | 2.675 | 0.271631263 |
| H17 | 2.209025512 | 2.35 | -0.140974488 |
| H18 | 1.873442614 | 1.65 | 0.223442614 |
| H20 | 3.483067617 | 3.65 | -0.166932383 |
| H21 | 3.559560436 | 3.78 | -0.220439564 |
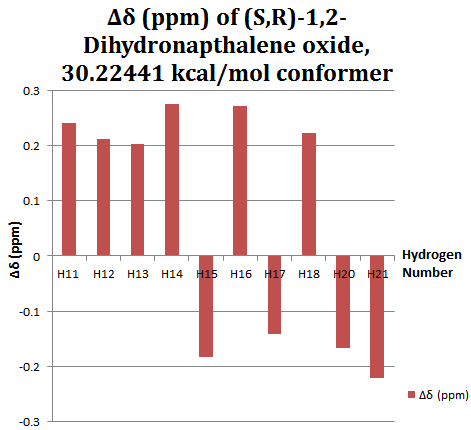
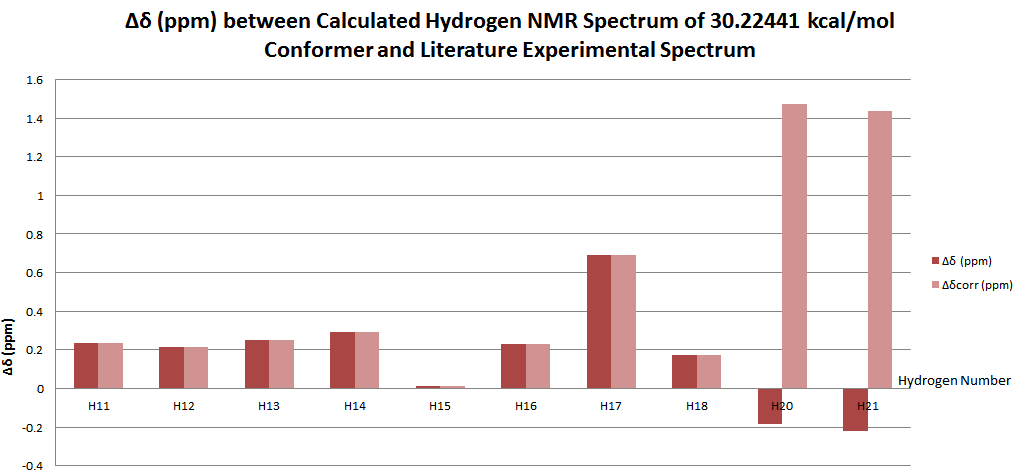
Carbon NMR Comparison with (-)-Rotamer
| Carbon Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| C1 | 29.06348904 | 20.8 | 8.263489036 | 0.03509053 | 8.263489036 | 8.263489036 | -4.120980016 | -22.37355 | |
| C2 | 52.82116189 | 54.1 | 31.72645 | -1.278838112 | -22.37355 | ||||
| C3 | 52.19244356 | 51.7 | 31.72645 | 0.492443564 | -19.97355 | ||||
| C4 | 130.3705995 | 131.5 | -1.129400532 | -1.129400532 | |||||
| C5 | 135.38776 | 135.6 | -0.212240025 | -0.212240025 | |||||
| C6 | 30.18028122 | 23.4 | 6.780281225 | 6.780281225 | |||||
| C7 | 123.533413 | 127.3 | -3.76658704 | -3.76658704 | |||||
| C8 | 123.7910881 | 127.4 | -3.60891185 | -3.60891185 | |||||
| C9 | 121.7441924 | 125.1 | -3.355807577 | -3.355807577 | |||||
| C10 | 126.6664766 | 128.5 | -1.833523393 | -1.833523393 | |||||
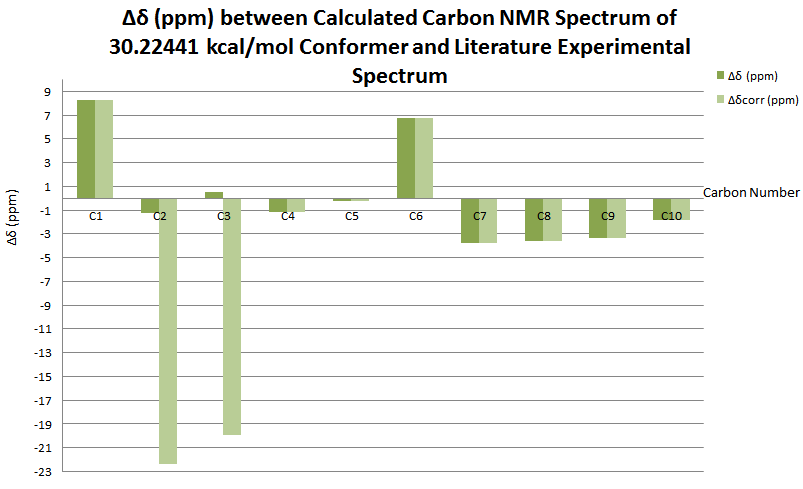
(-)-1,2-Dihydronapthalene oxide Literature Comparison
Further literature comparison with the (-)-rotamer was undertaken as a comparative study with the (+)-rotamer.
13C NMR data=
At the B3LYP 6-31G level of theory, 13C NMR spectrum is as below.
The Carbon shifts and disparity with literature[26] is as noted below.
| Carbon Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| C1 | 29.06348904 | 20.8 | 8.263489036 | 0.03509053 | 8.263489036 | 8.263489036 | -4.120980016 | -22.37355 | |
| C2 | 52.82116189 | 54.1 | 31.72645 | -1.278838112 | -22.37355 | ||||
| C3 | 52.19244356 | 51.7 | 31.72645 | 0.492443564 | -19.97355 | ||||
| C4 | 130.3705995 | 131.5 | -1.129400532 | -1.129400532 | |||||
| C5 | 135.38776 | 135.6 | -0.212240025 | -0.212240025 | |||||
| C6 | 30.18028122 | 23.4 | 6.780281225 | 6.780281225 | |||||
| C7 | 123.533413 | 127.3 | -3.76658704 | -3.76658704 | |||||
| C8 | 123.7910881 | 127.4 | -3.60891185 | -3.60891185 | |||||
| C9 | 121.7441924 | 125.1 | -3.355807577 | -3.355807577 | |||||
| C10 | 126.6664766 | 128.5 | -1.833523393 | -1.833523393 | |||||
The literature disparity is summarised in the following table.
Comparison of 13C NMR data with Literature
Repeat of NMR Analysis Using a Different Conformer and Higher Level of Theory
The MMFF94s force field, conjugate gradient algorithm optimised structures of the 30.2244 kcal/mol configurational isomer and the 30.6834 kcal/mol isomer were optimised further using Gaussian09 at the DFT B3LYP 6-31G(d,p) level of theory. These structures were input into optimisation at the higher DFT B3LYP cc-pVTZ level of theory, and NMR simulations carried out on both optimised structures. Results are as documented below.
30.68344 kcal/mol isomer at the 6-31G (d,p) level of theory
The 13C NMR spectrum of this isomer was corrected with reference TMS B3LYP/6-31G(d,p) Chloroform and reference shielding 192.17 ppm and NMR Degeneracy Tolerance of 0.05.
Atomic numbering used for the 30.68344 kcal/mol isomer is shown below.
The 13C NMR spectrum is shown below, with corrected shifts and comparison with literature,[26] tabulated.

| Carbon Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| C1 | 24.94531918 | 20.8 | 4.145319183 | -0.267577089 | -4.145319183 | 4.145319183 | 6.077712911 | 34.85481801 | |
| C2 | 56.4348396 | 54.1 | 88.1612896 | 2.334839596 | 34.0612896 | ||||
| C3 | 54.82836801 | 51.7 | 86.55481801 | 3.128368014 | 34.85481801 | ||||
| C4 | 130.383024 | 131.5 | -1.116975979 | -1.116975979 | |||||
| C5 | 133.8418328 | 135.6 | -1.758167178 | -1.758167178 | |||||
| C6 | 28.03616498 | 23.4 | 4.63616498 | 4.63616498 | |||||
| C7 | 123.8353838 | 127.3 | -3.464616185 | -3.464616185 | |||||
| C8 | 123.425036 | 127.4 | -3.974963965 | -3.974963965 | |||||
| C9 | 121.4413171 | 125.1 | -3.658682863 | -3.658682863 | |||||
| C10 | 125.5529435 | 128.5 | -2.947056492 | -2.947056492 | |||||

Applying the oxide C-O correction in fact increases the disparity with literature,[26] as can be seen in the following figure.

This may be due to the poor modelling of the simple ethylene oxide environment used for carbon correction factors when compared to the increased complexity of benzylic environment present for 1,2-dihydronapthalene. TALK ABOUT NAPTHALENE HERE!!!!
The equivalent analysis was carried out for the 1H NMR spectrum, and is documented below.

X-Axis: Shift (ppm)
- Y-Axis: Degeneracy
- Reference: TMS B3LYP/6-31G(d,p) Chloroform
- Reference shielding: 31.7462 ppm
- NMR Degeneracy Tolerance: 0.05
- X Y Degeneracy
14-H 7.4903000000 1.0000000000 1.0000000000 12-H 7.4035000000 1.0000000000 2.0000000000 13-H 7.3689000000 2.0000000000 2.0000000000 11-H 7.1952000000 1.0000000000 1.0000000000 21-H 3.6098000000 1.0000000000 2.0000000000 20-H 3.5657000000 2.0000000000 2.0000000000 16-H 2.8801000000 1.0000000000 1.0000000000 15-H 2.2677000000 1.0000000000 1.0000000000 18-H 2.2162000000 1.0000000000 1.0000000000 17-H 1.5628000000 1.0000000000 1.0000000000
To allow for comparison with literature, [27] the original spectra were used to refer to peaks notated as a "multiplet" in literature, and the peaks corresponding to one hydrogen environment were identified. For those hydrogens which observed coupling, each peak present in the multiplet was identified and these peaks averaged to find the centre of the multiplet, which corresponds to the chemical shift of the hydrogen.
| Hydrogen Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| H11 | 7.1952 | 7.018 | 0.1772 | 0.126956667 | 6.8408 | 0.1772 | 0.45889 | -0.050243333 | |
| H12 | 7.4035 | 7.1754 | 0.2281 | 0.2281 | |||||
| H13 | 7.3689 | 7.1356 | 0.2333 | 0.2333 | |||||
| H14 | 7.4903 | 7.322 | 0.1683 | 0.1683 | |||||
| H15 | 2.2677 | 2.2555 | 0.0122 | 0.0122 | |||||
| H16 | 2.8801 | 2.715666667 | 0.164433333 | 0.164433333 | |||||
| H17 | 1.5628 | 1.52 | 0.0428 | 0.0428 | |||||
| H18 | 2.2162 | 1.6988 | 0.5174 | 0.5174 | |||||
| H20 | 3.5657 | 3.667666667 | 5.225366667 | -0.101966667 | 1.5577 | ||||
| H21 | 3.6098 | 3.782 | 5.269466667 | -0.1722 | 1.487466667 | ||||
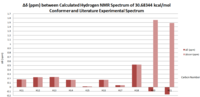
As for the carbon spectrum, applying the oxide hydrogen correction in fact increases the disparity between calculated results and literature, albeit the increase between these results is much lower, ~1.6 ppm for the hydrogen c.f ~32 ppm for the carbon NMR spectrum. This is most probably due to the increase distance between the hydrogens and epoxide oxygen, which gives the smaller correction factor calculated in the ethane model. Nevertheless, the increase in disparity reinforces the conclusion that the ethane environment is too oversimplified to allow for accurate correction factors. In general, the difference between the calculated and literature 1H NMR peaks[27] is lower than that observed for the 13C NMR spectrum.
30.68344 kcal/mol isomerAt the cc-pVTZ level of theory
H nmr spec napth oxide cc-pvtz 30.68344.PNG
Reference: TMS B3LYP/6-31G(d,p) Chloroform Reference shielding: 31.7462 ppm NMR Degeneracy Tolerance: 0.05
14-H 7.7291000000 1.0000000000 1.0000000000
12-H 7.5659000000 1.0000000000 2.0000000000
13-H 7.5307000000 2.0000000000 2.0000000000
11-H 7.4160000000 1.0000000000 1.0000000000
21-H 3.8126000000 1.0000000000 1.0000000000
20-H 3.6970000000 1.0000000000 1.0000000000
16-H 2.8681000000 1.0000000000 1.0000000000
15-H 2.5632000000 1.0000000000 1.0000000000
18-H 2.3767000000 1.0000000000 1.0000000000
17-H 1.6672000000 1.0000000000 1.0000000000
C nmr spec napth oxide cc-pvtz 30.68344.PNG
X-Axis: Shift (ppm)
- Y-Axis: Degeneracy
- Reference: TMS B3LYP/6-31G(d,p) Chloroform
- Reference shielding: 192.17 ppm
- NMR Degeneracy Tolerance: 0.05
- X Y Degeneracy
5-C 152.8919000000 1.0000000000 1.0000000000 4-C 148.8033000000 1.0000000000 1.0000000000
10-C 142.9270000000 1.0000000000 1.0000000000
8-C 141.3746000000 1.0000000000 1.0000000000 7-C 141.2615000000 1.0000000000 1.0000000000 9-C 139.0433000000 1.0000000000 1.0000000000 2-C 68.1034000000 1.0000000000 1.0000000000 3-C 65.4894000000 1.0000000000 1.0000000000 6-C 37.0983000000 1.0000000000 1.0000000000 1-C 34.1679000000 1.0000000000 1.0000000000
30.22440 kcal/mol isomer
C nmr spec napth oxide cc-pvtz 30.2244.PNG
X-Axis: Shift (ppm)
- Y-Axis: Degeneracy
- Reference: TMS B3LYP/6-31G(d,p) Chloroform
- Reference shielding: 192.17 ppm
- NMR Degeneracy Tolerance: 0.05
- X Y Degeneracy
5-C 152.8919000000 1.0000000000 1.0000000000 4-C 148.8034000000 1.0000000000 1.0000000000
10-C 142.9269000000 1.0000000000 1.0000000000
8-C 141.3746000000 1.0000000000 1.0000000000 7-C 141.2615000000 1.0000000000 1.0000000000 9-C 139.0433000000 1.0000000000 1.0000000000 2-C 68.1032000000 1.0000000000 1.0000000000 3-C 65.4893000000 1.0000000000 1.0000000000 6-C 37.0983000000 1.0000000000 1.0000000000 1-C 34.1679000000 1.0000000000 1.0000000000
H nmr spec napth oxide cc-pvtz 30.2244.PNG
X-Axis: Shift (ppm)
- Y-Axis: Degeneracy
- Reference: TMS B3LYP/6-31G(d,p) Chloroform
- Reference shielding: 31.7462 ppm
- NMR Degeneracy Tolerance: 0.05
- X Y Degeneracy
14-H 7.7291000000 1.0000000000 1.0000000000 12-H 7.5659000000 1.0000000000 2.0000000000 13-H 7.5307000000 2.0000000000 2.0000000000 11-H 7.4160000000 1.0000000000 1.0000000000 21-H 3.8126000000 1.0000000000 1.0000000000 20-H 3.6969000000 1.0000000000 1.0000000000 16-H 2.8681000000 1.0000000000 1.0000000000 15-H 2.5632000000 1.0000000000 1.0000000000 17-H 2.3767000000 1.0000000000 1.0000000000 18-H 1.6672000000 1.0000000000 1.0000000000
IR and VCD analysis
IR and VCD analysis was carried out for the lowest energy isomer at input 30.2244 kcal/mol, optimised to the DFT B3LYP cc-pVTZ level of theory, with results as below.
Napth ox 30.2244 IR cc-pVTZ im.PNG


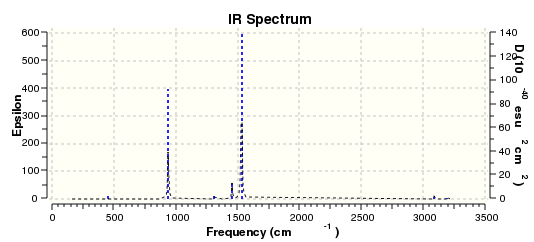
VCD can be used to elucidate the enantioeric excess (ee) of a sample consisting of a mixture of enantiomers of a chiral molecule. The sample % ee is defined as:
(NA - NB) x 100%/ (NA + NB)
where NA and NA is a measure of moles of enantiomer A and enantiomer B respectively. [28]This expression is equivalent to the excess amount of one enantiomer over the other relative to the total amount of both enantiomers. Optically pure samples of A have 100% ee, racemic mixtures 0% and optically pure B sample have -100% purity. VCD can be used for ee determination from the multiplicity of spectral bands that can be measure simultaneously in one spectrum. The intensity of the VCD bands gives an indication of the ee, as each VCD spectrum for each enantiomer is a mirror image of the other. [29]
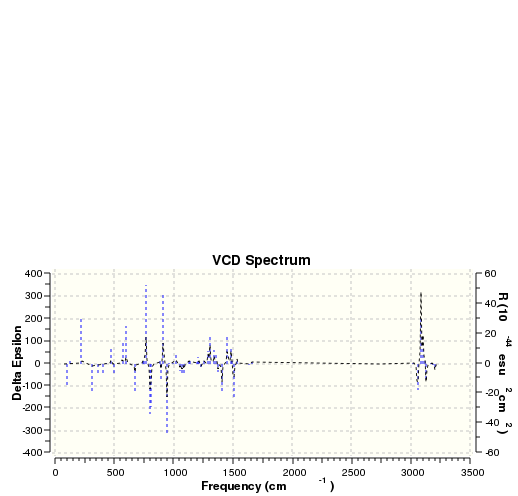
Styrene oxide Configurations
Styrene oxide has a chiral center at the benzylic carbon atom, giving (R)-styrene oxide and (S)-styrene oxide enantiomers. The energies of these two enantiomers were minimised to the MMFF94s, conjugate gradient level of theory.
| Enantiomer | Conformer 1 | Conformer 2 | Conformer 3 | |||
|---|---|---|---|---|---|---|
| Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | Structure | Energy Contribution (kcal/mol) | |
| R-styrene oxide | R-styrene oxide 21.61506 kcal/mol | Total Electronic Energy = 3.07365
Total Van der Waals = 13.67376 |
R-styrene oxide 21.61547 kcal/mol | Total Electronic Energy = 3.08594
Total Van der Waals = 21.61547 |
||
| S-styrene oxide | S-styrene oxide 21.61503 kcal/mol | Total Electronic Energy = 3.07620 Total Van der Waals = 13.67135 |
S-styrene oxide 21.61507 kcal/mol | Total Electronic Energy = 3.07301 Total Van der Waals = 13.67434 |
S-styrene oxide 21.61508 kcal/mol | Total Electronic Energy = 3.07293 Total Van der Waals = 13.67443 |
NMR analysis of S-styrene oxide
The NMR of (S)-styrene oxide was modelled at the DFT B3LYP 6-31G(d,p) level of theory in chloroform solvent, giving the structure as shown below.
(S)-styrene oxide at DFT B3LYP 6-31G(d,p)level of theory
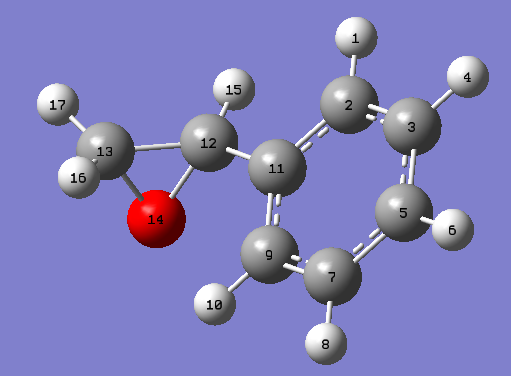
Following TMS/DFT B3LYP 6-31G(d,p) (chloroform) correction application, this gave 1H and 13C NMR spectra as documented in the figures below.
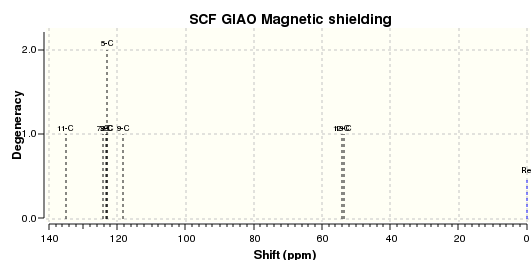
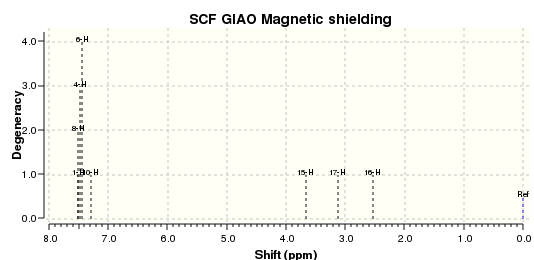
Numbering of atoms used in the following NMR analysis is demonstrated below.

Comparison of S-styrene Oxide with Literature
The carbon shifts were compared with both S-styrene oxide NMR and R-styrene oxide literature NMR shifts.
Comparison with R-styrene oxide literature shifts (K. Sarma, N. Bhati et al.) is as documented below.
| Carbon Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| C2 | 122.9659755 | 128.1 | -5.134024522 | -7.231939222 | 5.134024522 | 11.7736977 | 33.982242 | |
| C3 | 123.4152252 | 128.5 | -5.0847748 | 5.0847748 | ||||
| C5 | 122.9534464 | 125.5 | -2.546553554 | 2.546553554 | ||||
| C7 | 124.1326453 | 128.5 | -4.367354719 | 4.367354719 | ||||
| C9 | 118.2680608 | 125.5 | -7.231939222 | 7.231939222 | ||||
| C11 | 135.1343367 | 137.6 | -2.465663325 | 2.465663325 | ||||
| C12 | 54.05057945 | 52.4 | 85.77702945 | 1.650579448 | 33.37702945 | |||
| C13 | 53.455792 | 51.2 | 85.182242 | 2.255791999 | 33.982242 | |||
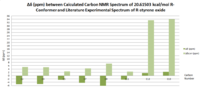
Comparison with S-styrene oxide NMR literature was carried out,(Synthesis 2011(7): 1092-1098) with results as follows:
13C NMR
| Carbon Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| C2 | 122.9659755 | 128.1 | -5.134024522 | -2.865492337 | -7.231939222 | 5.134024522 | 11.7736977 | 33.982242 | |
| C3 | 123.4152252 | 128.5 | -5.0847748 | 5.0847748 | |||||
| C5 | 122.9534464 | 125.5 | -2.546553554 | 2.546553554 | |||||
| C7 | 124.1326453 | 128.5 | -4.367354719 | 4.367354719 | |||||
| C9 | 118.2680608 | 125.5 | -7.231939222 | 7.231939222 | |||||
| C11 | 135.1343367 | 137.6 | -2.465663325 | 2.465663325 | |||||
| C12 | 54.05057945 | 52.4 | 85.77702945 | 1.650579448 | 33.37702945 | ||||
| C13 | 53.455792 | 51.2 | 85.182242 | 2.255791999 | 33.982242 |
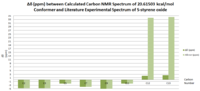
Theoretically, no difference should be observed between the experimental NMR of the R- and S- enantiomers. However, comparing the carbon NMR shifts obtained through B3LYP 6-311G(d,p) optimisation and simulation, the shift differences of the carbon NMR spectra with literature reveal that a closer fit is obtained with the literature S-styrene oxide NMR. (Synthesis) The disparity shown between the calculated and experimental shifts is indicative of the conformational transitions and intermolecular coupling [1] in solution that are not modelled computationally.
1H NMR
The hydrogen shifts were compared with literature experimental spectra. The S-isomer literature spectrum [30] when compared to the calculate R-isomer spectrum gave parameters as shown.
| Hydrogen Number | δcalc (ppm) | Multiplet δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| H1 | 7.514585625 | 7.451002894 | 7.28 | 0.171002894 | -0.069801232 | -0.251973718 | 0.171002894 | 1.174948768 | 1.644740884 | |
| H4 | 7.483516459 | |||||||||
| H6 | 7.446452754 | |||||||||
| H8 | 7.512661831 | |||||||||
| H10 | 7.2977978 | |||||||||
| H15 | 3.662441679 | 3.84575 | 5.322108346 | -0.183308321 | 1.476358346 | |||||
| H16 | 2.534026282 | 2.786 | 4.193692949 | -0.251973718 | 1.407692949 | |||||
| H17 | 3.114074217 | 3.129 | 4.773740884 | -0.014925783 | 1.644740884 |
This is summarised in the shift difference graph below.
![S-Styrene oxide hydrogen NMR shift difference graph with literature[30]](/images/thumb/0/0a/S-STYRENE_EPOXIDE_COMPARISON_WITH_HYDROGEN_S-STYRENE_EPOXIDE_LITER_DIFF_TABLE_6-31G.PNG/200px-S-STYRENE_EPOXIDE_COMPARISON_WITH_HYDROGEN_S-STYRENE_EPOXIDE_LITER_DIFF_TABLE_6-31G.PNG)
Comparison with R-styrene oxide literature (K. Sarma, N. Bhati) gave a slightly closer fit of calculated hydrogen shifts to experimental shifts.
| Hydrogen Number | δcalc (ppm) | Multiplet δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| H1 | 7.514585625 | 7.451002894 | 7.31 | 0.141002894 | -0.094613732 | -0.275973718 | 0.141002894 | 1.150136268 | 1.623740884 | |
| H4 | 7.483516459 | |||||||||
| H6 | 7.446452754 | |||||||||
| H8 | 7.512661831 | |||||||||
| H10 | 7.2977978 | |||||||||
| H15 | 3.662441679 | 3.87 | 5.322108346 | -0.207558321 | 1.452108346 | |||||
| H16 | 2.534026282 | 2.81 | 4.193692949 | -0.275973718 | 1.383692949 | |||||
| H17 | 3.114074217 | 3.15 | 4.773740884 | -0.035925783 | 1.623740884 |
The closer fit is summarised in the shift difference graph below.
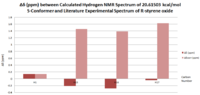
The closer fit of carbon NMR to the S-Styrene oxide literature conflicts with the closer fit of the hydrogen NMR to the R-Styene oxide literature, hinting towards the theoretical equivalence of the R- and S-enantiomer NMR spectra. Further literature would need to be carried out to gain a wider appreciation of such an effect. Additionally, it can be seen that correcting for the oxide hydrogens and carbons in fact increases disparity with literature.
NMR analysis of R-Styrene Oxide Lowest Energy Conformer
The 20.61505 kcal/mol conformer was optimised using the DFT B3LYP 6-31G(d,p) level of theory in Gaussian09 and NMR simulation calculatioins were carried out. This gave NMR spectra as shown below.

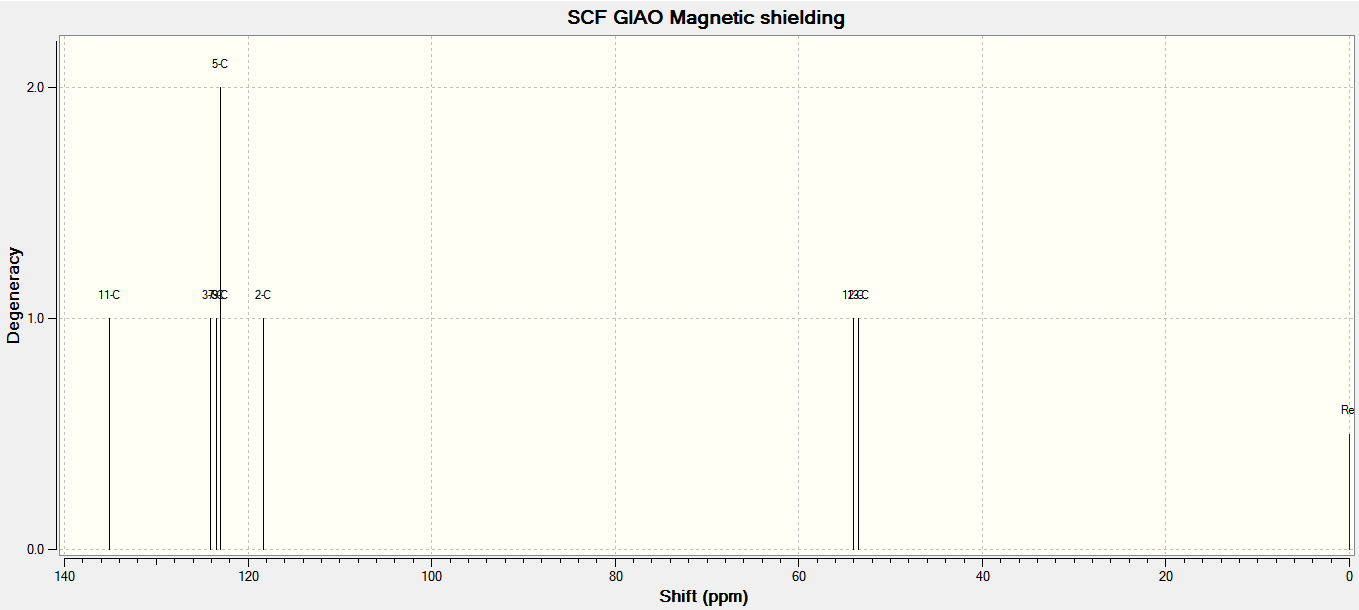
Atomic numbering used when referring to specified atoms in NMR analysis is as shown below.
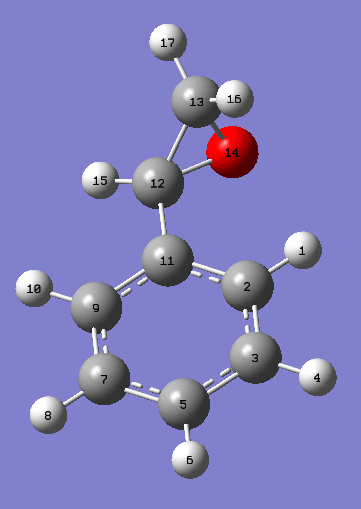
Comparison of spectra with (S)-Styrene oxide
Literature NMR spectra of S-Styrene oxide, [31] was compared with the calculated NMR spectra. Results for 13C NMR spectra are as shown below in the following table and shift difference graph.
| Carbon Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| C2 | 118.2706444 | 127.7 | -9.429355626 | -2.463345741 | -9.429355626 | -9.429355626 | 1.502460509 | 2.573722263 | |
| C3 | 124.1340577 | 128.1 | -3.965942316 | -3.965942316 | |||||
| C5 | 122.9525029 | 125.1 | -2.147497079 | -2.147497079 | |||||
| C7 | 123.4136518 | 128.1 | -4.686348199 | -4.686348199 | |||||
| C9 | 122.9586151 | 125.1 | -2.141384907 | -2.141384907 | |||||
| C11 | 135.1341726 | 137.2 | -2.065827374 | -2.065827374 | |||||
| C12 | 54.05586731 | 51.9 | 85.78231731 | 2.155867307 | 33.88231731 | ||||
| C13 | 53.47372226 | 50.9 | 85.20017226 | 2.573722263 | 2.573722263 |
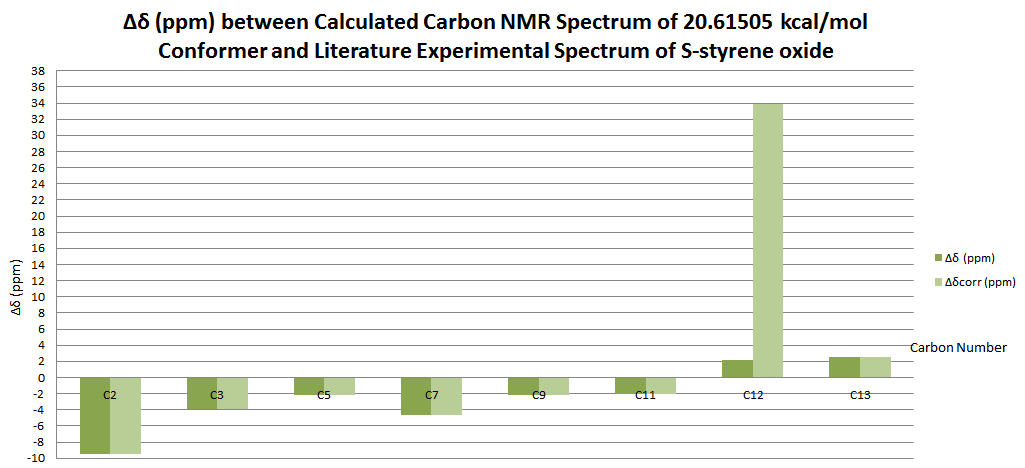
Comparison of spectra with (R)-Styrene oxide
Literature NMR spectra of R-Styrene oxide,[32] was compared with the calculated NMR spectra. Results for 13C NMR spectra are as shown below in the following table and shift difference graph.
| Carbon Number | δcalc (ppm) | δexp (ppm) | δcorr (ppm) | ∆δ (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) | ∆δcorr (ppm) | Average ∆δ (ppm) | Mode ∆δ (ppm) |
| C2 | 118.2706444 | 128.1 | -9.829355626 | -2.863345741 | -9.829355626 | -9.829355626 | 5.068266759 | 34.00017226 | |
| C3 | 124.1340577 | 128.5 | -4.365942316 | -4.365942316 | |||||
| C5 | 122.9525029 | 125.5 | -2.547497079 | -2.547497079 | |||||
| C7 | 123.4136518 | 128.5 | -5.086348199 | -5.086348199 | |||||
| C9 | 122.9586151 | 125.5 | -2.541384907 | -2.541384907 | |||||
| C11 | 135.1341726 | 137.6 | -2.465827374 | -2.465827374 | |||||
| C12 | 54.05586731 | 52.4 | 85.78231731 | 1.655867307 | 33.38231731 | ||||
| C13 | 53.47372226 | 51.2 | 85.20017226 | 2.273722263 | 34.00017226 |
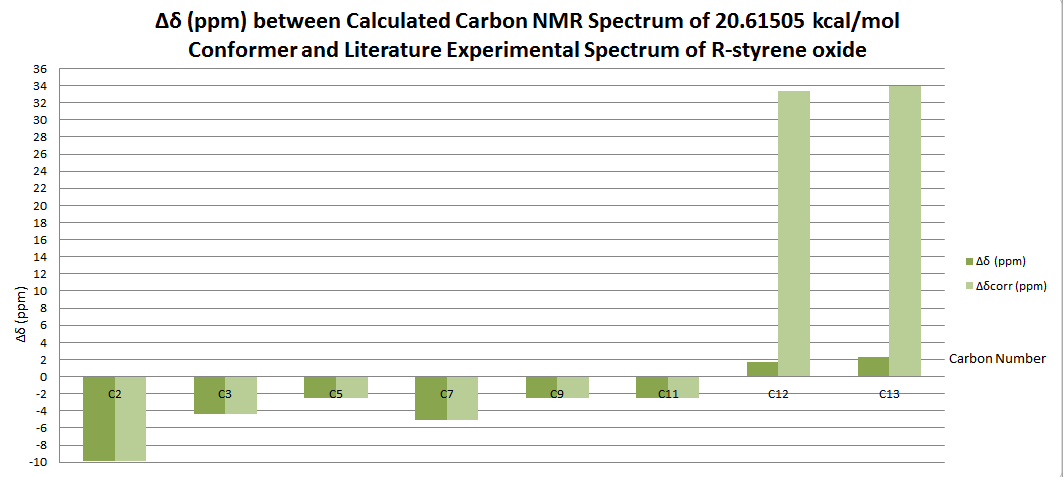
IR and VCD analysis of styrene oxide
S-Styrene Oxide
The IR peaks of the lowest energy S-styrene oxide enantiomer, of energy 20.61503 kcal/mol, were compared with literature,[1] with results tabulated below.
| ν (cm-1 | εcalc | ∆νcalc-exp | Average ∆ν | Mode ∆ν |
|---|---|---|---|---|
| 3031 | 0.8672364229 | -4 | 0.785714286 | 12 |
| 3038 | 1.1258300005 | 3 | ||
| 2975 | 0.2298337644 | 0 | ||
| 2919 | 0 | -2 | ||
| 2926 | 0 | 5 | ||
| 2933 | 0.0777506685 | 12 | ||
| 1603 | 0.3872326267 | -2 | ||
| 1456 | 2.1668723937 | 1 | ||
| 1281 | 5.3363537201 | -5 | ||
| 1288 | 24.6060915808 | 2 | ||
| 980 | 2.5214957845 | -1 | ||
| 875 | 6.9168187410 | 2 | ||
| 763 | 35.0258886184 | 3 | ||
| 693 | 3.9809019461 | -3 |
=R-Styrene Oxide
The IR spectrum for the R-styrene oxide lowest energy isomer was modelled at the DFT B3LYPP 6-31G(d,p) level of theory, and is as shown below.
Through infrared spectroscopy, molecules absorb specific frequencies of non-polarised infrared red that are characteristic of their bonding structure, so creating normal modes with resonant frequencies related to bond strength and atomic mass of the atoms involved in bonding. For non-linear molecules like styrene-oxide, the degrees of vibrational freedom are 3N-6, where N signifies the number of atoms in the molecule, Thus, as IR spectra are indicative of bonding in the molecule, it is well documented that configurational enantiomers are indistinguishable by IR spectroscopy, due to no change in bonding structure occurring between them. However, the R-styrene oxide calculated spectrum is somewhat offset from the calculated S-styrene oxide spectrum and experimental literature spectra.[1] [1]
This disparity occurs most dramatically in the 2700-3000 cm-1 region, in which the R-isomer has an absence of characteristic peaks. This is indicative of the static nature of the calculation, which takes into account only the one supplied conformation of the configurational isomer being investigated. In reality, the experimental spectrum occurs with rapid interchange between conformers of the configuration. Thus the total bond energies are averaged over a selection of conformations, and the total bond energies of both R and S-styrene oxide have equivalence. Additionally, this disparity between IR spectra may be indicative of a poor R- or S-styrene model used as input for the NMR calculation.
VCD of S-enantiomer
The VCD of the S-enantiomer was run at the DFT B3LYP 6311++G(2df,p) level of theory also, giving a spectrum as shown.
As reported by G.-J. Kim, D.-W. Park et al.[29] the VCD of S-(-)-styrene oxide, at the maximum intensity, gives a positive intensity at 1250 cm-1, whilst R-(+)-styrene oxide gives a negative intensity of equal magnitude at equivalent frequencies. Thus, as the assigned S-enantiomer gives a maximum positive intensity in the present results, some agreement with literature is observed.[29] However, the maximum peak, lying at 1500 cm-1 is removed by ~250 cm-1 from the expected maximum peak, albeit the reduced intensities in the 1200-1000 cm-1 region agree well with literature.[29]
VCD of R-enantiomer
The VCD of the R-enantiomer was run at the DFT B3LYP 6311++G(2df,p) level of theory also, giving a spectrum as shown.

As reported by G.-J. Kim, D.-W. Park et al.[29] the VCD of R-(+)-styrene oxide, at the maximum intensity, gives a negative intensity at 1250 cm-1, whilst S-(+)-styrene oxide gives a positive intensity of equal magnitude at equivalent frequencies. Thus, as the assigned R-enantiomer gives a maximum positive intensity in the present results, some agreement with literature is observed.[29] However, the maximum peak, lying at 1500 cm-1 is removed by ~250 cm-1 from the expected maximum peak, albeit the reduced intensities in the 1200-1000 cm-1 region agree well with literature.[29]
Optical Rotations, ECD and UV-Vis of the Epoxides
For conformational isomers, stereochemistry is only important if one conformation cannot inter-convert freely with the other. Purification of one member of the enantiomeric pair or chiral diastereomers must be achieved to observe an optical rotation; members of the pair cancel each other out if purification is not achieved. Enantiomers of a known compound have optical rotations opposite in sign but identical in intensity. However it is not possible to predict the sign or magnitude of an enantiomer from its stereochemistry. Two diastereomers however have no reported relationship with respect to optical activity sign or magnitude, and have differences in other characteristic properties e.g. reactivity, melting point and solubility that cannot be predicted. For NMR and IR, enantiomers have the same spectra, but diastereomers have different spectra.
Optical Rotation of 1,2-Dihydronapthalene Oxide
The optical rotation of the 1,2-Dihydronapthalene oxide structure can be found from either the 1R,2S- or 1S,2R-enantiomer, as each is of the same magnitude. The lowest energy conformer of each configuration provides the most accurate optical rotation, at room temperature, of that configuration, as most time is spent in the lowest energy conformation, and so at any one time in a given sample, the dominant conformation exists at the energy minimum. The optical rotation was trialled for a number of isomers in order to check reliability of results. The higher energy (1S,2R)-isomer, of 30.68344 kcal/mol, was optimised to the DFT B3LYP 6-31G(d,p) level of theory, the optical rotation was calculated with the 6,311++G(2df,p) basis set. The 30.68344 kcal/mol isomer gave a negative rotation, of magnitude shown below.
[Alpha] ( 5890.0 A) = -155.82⁰
[Alpha] ( 3650.0 A) = -522.16⁰
The 30.2244 kcal/mol isomer, the lowest energy (1S,2R)-isomer, and thus the most accurate approximation for optical rotation was optimised to the DFT B3LYP 6-31G(d,p) level of theory and the optical rotation was calculated with the 6,311++G(2df,p) basis set, giving a negative rotation of magnitude shown below.
[Alpha] ( 5890.0 A) = -155.82⁰
[Alpha] ( 3650.0 A) = -522.15⁰
To increase accuracy of this calculation further the structure was optimised further using the higher force field cc-pVTZ and optical rotation simulation carried out. This returned reduced o.r as below.
[Alpha] ( 5890.0 A) = -155.22⁰
[Alpha] ( 3650.0 A) = -516.50⁰
The negative rotation for the 1S,2R-isomer compares favourably with literature reports,[33] but indicates the %ee assignment in such literature may be drawn to error due to contaminant molecules present in so-called "enantiopure" references. For example, S. Pedragosa-Moreau et al.[33] reported a 99% ee with o.r +129 for the (1R,2S)-isomer. This result implies a 100% enantiopure substance of o.r 130.303⁰, a value which is 25.17⁰ and 24.92⁰ lower than that calculated using the 30.2244 kcal/mol isomer, with input optimisations of DFT B3LYP 6-31G(d,p) and cc-pVTZ level of theory respectively. Indeed, the higher energy 30.68344 kcal/mol input isomer at optimised to B3LYP 6-31G(d,p) level of theory also returns an enantiopure o.r greater than the experimental reference. As calculations on the lowest energy isomer at both levels of theory return an enantiopure optical rotation greater in magnitude than the experimental enantiopure reference, this may indicate that the true enantiopure o.r lies significantly higher than the 130.303⁰ reported. However, optical activity is not an individual molecular property, but results from the random distribution of sample molecules. The calculation performed presently is performed on one static molecule only, and does not take into account inter-molecular and packing effects, which may cause changes to the molecular geometry of the oxides and thus cause deviations from the single molecule enantiopure o.r. Additionally, o.r measurements are sensitive to temperature fluctuations, and any disparity between the calculation parameters and the environment of the enantiopure substance may cause the observed experimental o.r reduction.
Optical Rotation of Styrene Oxide
Optical Rotation of S-(-)-Styrene Oxide
Using the NMR of the S-Styrene oxide, the lowest energy structure found in the present study, optimised to the DFT B3LYP 6-31G(d,p) level of theory, the optical rotation was calculated with the 6,311++G(2df,p) basis set. In accordance with literature,[34] [35]this gave a negative rotation for S-styrene oxide, of magnitude shown below.
[Alpha] ( 589.0 nm) = -30.07⁰
[Alpha] ( 365.0 nm) = -93.90⁰
This o.r compared poorly with literature values of ~23⁰,[34] [35] and so the enantiomer was optimised further to the cc-pVTZ level of theory. Using the NMR of this S-Styrene oxide optimised to the DFT B3LYP cc-pVTZ level of theory, the optical rotation was calculated with the 6,311++G(2df,p) basis set. The S-conformer gave a negative rotation, of magnitude shown below.
[Alpha] ( 589.0 nm) = -22.52⁰
Alpha] ( 365.0 nm) = -69.26⁰
The optical rotation at the higher level of theory agrees well with a -22.5⁰ enantiomeric pure reference in literature at 26⁰C[34], but differs by 1.42⁰ from reports by D. E. White et al. with an enantiomeric pure reference of -23.94⁰ at a temperature of 31⁰C.[34] [36] These differences may arise from temperature differences at measurement of the optical rotation but may also arise from an enantiomeric contamination present for the higher optical rotation. The present optical rotation is reduced compared to those obtained in previous reports due to the averaging effects of conformational transitions in solution, which are not accounted for by the static conformer model. Experimentally, this results in an average optical rotation of the conformations involved in conformational transitions. The present calculation has taken into account only one conformation. To reduce the effects of conformational transitions during computational analysis, the lowest energy conformer found for the S-Styrene oxide product was input into the optical rotation analysis as, this being the lowest energy conformer found presently, it was expected to exist predominantly in solution at any given time, and so give the greatest contribution to the optical rotation. However, the conformation found presently may reside at a local energy minima, in which case the trialed conformer would not be present as the predominant conformation.
Optical Rotation of R-(+)-Styrene Oxide
Using the NMR of R-Styrene oxide optimised to the DFT B3LYP 6-31G(d,p) level of theory, the optical rotation was calculated with the 6.311++G(2df,p) basis set. The higher energy 21.61547 kcal/mol configuration gave a positive rotation, of magnitude shown below.
[Alpha] ( 589.0 A) = 30.17⁰ [Alpha] ( 365.0 A) = 94.24⁰
Using the NMR of Styrene oxide optimised to the DFT B3LYP cc-pVTZ level of theory, the optical rotation was calculated with the 6,311++G(2df,p) basis set. The higher energy 21.61547 kcal/mol configuration gave a positive rotation, of magnitude shown below.
[Alpha] ( 589.0 A) = 22.47⁰ [Alpha] ( 365.0 A) = 69.10⁰
The R-styrene oxide conformer found with lower energy, 21.61506 kcal/mol was optimised to the DFT B3LYP 6-31G(d,p) level of theory, the optical rotation was calculated with 6.311++G(2df,p) basis set.
[Alpha] ( 589.0 A) = 30.43⁰ [Alpha] ( 365.0 A) = 95.05⁰
This conformer was optimised to the DFT B3LYP cc-pVTZ level of theory, and the optical rotation was calculated with 6.311++G(2df,p) basis set, giving a closer fit with literature.[37]
[Alpha] ( 589.0 A) = 23.06⁰ [Alpha] ( 365.0 A) = 71.02⁰
In general for the Styrene oxide enantiomers, the lower energy conformers give the best fit with literature, and increasing the optimisation level of theory to the cc-pVTZ level increases the agreement with literature.[37]
ECD and UV-Vis Simulations
Electronic Circular Dichroism spectroscopy (ECD) can be used to determine the absolute structure of solid and solution-phase molecules. For determining molecular 3D structure, configuration and conformation, it is a sensitive, quick and reliable technique, provided that the substance has chirality, is non-racemic, and absorbs circularly polarized light (CPL) in 180 - 1100 nm range available.[38]
ECD and UV-Vis of 1,2-Dihydronapthalene Oxide
ECD and UV-Vis simulation were run on the 30.22441 kcal/mol configuraiton isomer, at DFT B3LYP 6-31G level of theory giving spectra as shown below.
- UV-vis napthalane lowest energy conformer.svg
Excitation Energy (nm) = 192.5000000000 ɛ = 57629.5162835934 d(epsilon)/d(excitation energy) = -294.5415960797
- Ecd spectrum napthalene lowest energy.svg
Excitation Energy (nm)= 196.0000000000 ∆ɛ = 9.0192453436
ECD and UV-Vis simulations were also run on the lowest energy conformer, the 30.2244 kcal/mol of the (-)-1,2-dihydronapthalene oxide configuration, which was deemed to be a more accurate representation of the molecule in solution. Noted are the maxima and minima for the relevant spectra of the structure optimised to the B3LYP 6-31G(d,p) level of theory are as below.
- UV-Vis spectrum:
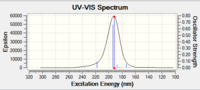
Excitation Energy (nm)= 192.02
Oscillator Strength = 0.7778
Comparing this UV-Vis maximum with that obtained for the higher energy (+)-isomer at the same level of theory, there is a 0.48 nm decrease in absorption maximum associated with the 0.00001 kcal/mol decrease in initial input energy.
- ECD spectrum:
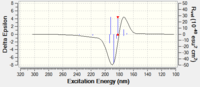
Excitation Energy (nm)= 187.58
Rvel (10-40 esu2cm2 = -47.1
∆ɛ = ~-8
Excitation Energy (nm)= 173.5
Rvel (10-40 esu2cm2 = 26.872
∆ɛ = ~4.5
To increase the accuracy of these calculations, the structure was optimised to B3LYP cc-pVTZ level of theory and ECD and UV-Vis simulations run. Noted are the maxima and minima for the relevant spectra of the structure optimised to the cc-pVTZ level of theory are as below.
- UV-Vis spectrum:
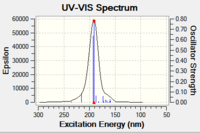
Excitation Energy (nm)= 190.84
Oscillator Strength = 0.7719
Thus increasing the force field increases the energy of the maximum absorption excitation energy, which is to be expected given the reduction in total energy that the cc-pVTZ force field brings about during optimisation c.f. the 6-31G force field.
- ECD spectrum:
Excitation Energy (nm)= 158.78
Rvel (10-40 esu2cm2 = 0.441
∆ɛ = ~-3
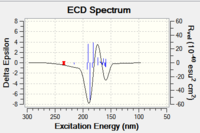
Excitation Energy (nm)= 173.53
Rvel (10-40 esu2cm2 = 7.1378
∆ɛ = ~3
Excitation Energy (nm)= 187.18
Rvel (10-40 esu2cm2 = -530.165
∆ɛ = ~-8
At the higher level of theory, one extra peak at a higher energy of 158.78 nm is observed, and all peaks are shifted to lower energy.
The ECD spectra are in general the mirror image of the UV-Vis spectrum, as expected, and each configurational isomer ECD spectrum is the mirror image of the other. However, no literature could be found for either enantiomer to compare with experimental values. Literature reports involving napthalene derivatives exhibit ECD peaks within the 200-250 nm region.[39] The calculated values at the DFT B3LYP 6-31G level of theory lie in the 150-200 nm region, and thus provide a reasonable similarity given the errors associated with such a comparison.
ECD and UV-Vis of Styrene Oxide
Using the NMR calculation carried out on the R-styrene oxide highest energy configurational isomer, at the DFT B3LYP 6-31G(d,p) level of theory, ECD and UV-Vis simulations were run, giving results as shown below.
- UV-Vis Spectrum

ʎmax Excitation Energy (nm) = 187.98
Oscillator Strenght = 0.867
∆ɛ = ~59000
- ECD Spectrum

ʎmax Excitation Energy (nm)= 185.54
∆ɛ = ~-12
Rvel = ~-30
ʎmax Excitation Energy (nm)= 160.09
∆ɛ = ~15
Rvel = ~40
Using the NMR calculation carried out on the S-styrene oxide configurational isomer, of lowest energy, at the DFT B3LYP 6-31G(d,p) level of theory, ECD and UV-Vis simulations were run, giving results as shown below.
- UV-Vis Spectrum

ʎmax Excitation Energy (nm) = 188.2400000000
∆ɛ = 58894.6631990984
d∆ɛ/dExcitation Energy =320.5154439262
- ECD Spectrum

ʎmax Excitation Energy (nm)= 185.6000000000
∆ɛ = 13.8016611316
d∆ɛ/dExcitation Energy = -0.4784170592
ʎmin Excitation Energy (nm) = 160.0800000000
∆ɛ = -15.8501295345
d∆ɛ/dExcitation Energy = 0.3810660432
Suggesting New Candidates for Investigation=
A suitable epoxide of o.r > 500 or < 500 is provided by 2-(4-methylphenyl)-3-phenyl oxirane trans-(2R,3R)-2-(4-methylphenyl)-3-phenyl oxirane [Alpha] ( 5890.0 A) = -320.35˚ [Alpha] ( 4360.0 A) = -730.74˚
cis [Alpha] ( 5890.0 A) = -25.48 deg [Alpha] ( 4360.0 A) = -49.25 deg

Dansette,P.M. et al. Tetrahedron, 1976 , vol. 32, p. 2071 - 2074
The trans-(2R,3R)-2-(4-methylphenyl)-3-phenyl oxirane was optimised to a minimum in Avogadro using the MMF94s force field and conjugate gradients. The structure and parameters of the optimised molecule are shown below.
Trans methyl stilbene for hpc.cml
| Energy Contribution | Energy (kcal/mol) |
| Total Electrostatic Energy | 5.04177 |
| Total Out-of-Palne Bending Energy | 0.0201 |
| Total Torsional Energy | 2.17991 |
| Total Stretch Bending Energy | -1.50356 |
| Total Angle Bending Energy | 2.83423 |
| Total Bond Stretching Energy | 3.9429 |
| Total Energy | 41.55575 |
This structure was submitted to optimisation at the DFT B3LYP 6-31G(d,p) level of theory, giving a structure and energy as shown below.
Sum of electronic and zero-point Energies= -655.026348
Sum of electronic and thermal Energies= -655.012762 Sum of electronic and thermal Enthalpies= -655.011818 Sum of electronic and thermal Free Energies= -655.068821
The enantiomeric (1S,2R)-cis-2-(4-methylbenzenyl)-3-phenyl-oxirane, with a lower reported optical rotation, was optimised in Avogadro with the MMFF94s force field using the conjugate gradient algorithm. The structure and parameters of the lowest energy structure obtained using this technique are as below.
Trans methyl stilbene opt 4 (cis) new one.cml
| ' | |
| Energy Contribution | Energy (kcal/mol) |
| Total Electrostatic Energy | 6.56941 |
| Total Van der Waals Energy | 28.65841 |
| Total Out-of-Palne Bending Energy | 0.02091 |
| Total Torsional Energy | 2.1782 |
| Total Stretch Bending Energy | -1.3851 |
| Total Angle Bending Energy | 3.20829 |
| Total Bond Stretching Energy | 4.08582 |
| Total Energy | 43.33594 |
NMR analysis was carried out, giving good agreement with literature for most carbon shifts, as documented below.
| Carbon Number | δcalc (ppm) | δexp (ppm) | ∆δcalc-exp (ppm) | Average ∆δcalc-exp (ppm) | Mode ∆δcalc-exp (ppm) |
| C1 | 122.9778008 | 125.5 | -2.522199225 | -4.017924319 | -7.153745456 |
| C2 | 123.513071 | 128.5 | -4.986928969 | ||
| C3 | 123.0827678 | 128.2 | -5.117232162 | ||
| C4 | 124.1918224 | 129.2 | -5.008177643 | ||
| C5 | 118.2462545 | 125.4 | -7.153745456 | ||
| C6 | 134.2028056 | 138.2 | -3.997194358 | ||
| C7 | 124.2666619 | 129.2 | -4.933338102 | ||
| C8 | 123.100 | 128.2 | -5.100009147 | ||
| C9 | 131.2212506 | 138.2 | -6.978749362 | ||
| C10 | 118.4044517 | 125.4 | -6.99554825 | ||
| C11 | 124.9491655 | 137.2 | -12.25083451 | ||
| C12 | 134.7213106 | 138.2 | -3.478689364 | ||
| C13 | 66.42459374 | 62.8 | 3.624593736 | ||
| C14 | 66.27088104 | 62.7 | 3.57088104 | ||
| C16 | 22.25830698 | 21.2 | 1.058306982 | ||
This was optimised to the DFT B3LYP 6-31G(d,p) level of theory, giving a structure of energetic parameters, expressed in Hartree/particle, as below.
Thermal correction to Gibbs Free Energy= 0.204857
Sum of electronic and zero-point Energies= -655.023912
Sum of electronic and thermal Energies= -655.010405
Sum of electronic and thermal Enthalpies= -655.009460
Sum of electronic and thermal Free Energies= -655.066618
- ↑ 1.0 1.1 1.2 1.3 1.4 1.5 1.6 W. T. Klooster , T. F. Koetzle , P. E. M. Siegbahn , T. B. Richardson , and R. H. Crabtree, "Study of the N-H···H-B Dihydrogen Bond Including the Crystal Structure of BH3NH3 by Neutron Diffraction", J. Am. Chem. Soc., 1999, 121, 6337–6343.DOI:10.1021/ja9825332 Cite error: Invalid
<ref>tag; name "ja9825332" defined multiple times with different content - ↑ 2.0 2.1 2.2 J. Sauer, R. Sustmann, Angew. Chem. Int. Ed. Engl., 1980, 19, 779 - 807.DOI:[1] Cite error: Invalid
<ref>tag; name "ja9825334" defined multiple times with different content - ↑ K. Alder, G. Stein , Angew. Chem., 1937, 50, 510.DOI:[2]
- ↑ L. Paquette, N. A. Pegg, D. Toops, G. D. Maynard, R. D. Rogers, J. Am. Chem. Soc., 1990, 112, 277-283.DOI:10.1021/ja9825332
- ↑ 5.0 5.1 5.2 5.3 S. W. Elmore, L. Paquette, Tet. Lett., 1991, 319, 1891DOI:10.1016/S0040-4039(00)92617-0
- ↑ 6.0 6.1 W. F. Maier, P.-V. R. Schleyer,J. Am. Chem. Soc., 1981, 103, 1891DOI:10.1021/ja00398a003
- ↑ C. Braddock, H. S. Rzepa, J. Nat. Prod., 2008, 71, 728-730DOI:10.1021/np0705918
- ↑ M. Durík, V. Langer, D. Gyepesová, J. Micová, B. Steiner M. Koós, Acta Cryst.,2001,E57, 672-674DOI:10.1107/S160053680101073X 10.1107/S160053680101073X
- ↑ 9.0 9.1 Z.-X. Wang , S. M. Miller, O. P. Anderson, Y. Shi, J. Org. Chem.,2001, 66, 2, 521–530.DOI:10.1021/jo001343i 10.1021/jo001343i
- ↑ 10.0 10.1 10.2 R. V. Stick, S. J. Williams, Carbohydrates, Amsterdam, Elsevier, 2009.Template:ISBN Cite error: Invalid
<ref>tag; name "ja9825335" defined multiple times with different content - ↑ 11.0 11.1 11.2 J. Clayden, N. Greeves, S. Warren, P. Wothers, Organic Chemistry, Oxford University Press,2nd Edition, 2012, 1126-1131.Template:ISBN
- ↑ Z.-X. Wang , S. M. Miller, O. P. Anderson, Y. Shi, J. Org. Chem.,2001, 66, 2, 521–530.DOI:10.1021/jo001343i 10.1021/jo001343i
- ↑ J. W. Yoon, T.-S. Yoon, S. W. Lee, W. Shin, Acta Cryst.,1999, C55, 1766-1769 .DOI:10.1107/S0108270199009397
- ↑ F. R. Fronczek, X. R. J. Bu, Private Comm.,2002DOI:[3]
- ↑ 15.0 15.1 E. Anslyn, Modern Physical Organic Chemistry, 2004, Sausalito, CA, University ScienceTemplate:ISBN Cite error: Invalid
<ref>tag; name "ja9825332123" defined multiple times with different content - ↑ Z.-X. Wang,Y. Shi, J. Org. Chem. 1997, 62, 8622-8623DOI:S0022-3263(97)01701-5
- ↑ 17.0 17.1 17.2 17.3 G.-A. Cao, Z.-X. Wang, Y. Tu, Y. Shi, Tetrahedron Letters, 1998, 39, 25, 4425–4428DOI:10.1016/S0040-4039(98)00838-7 Cite error: Invalid
<ref>tag; name "ja982533672123" defined multiple times with different content - ↑ M. Hickey, D. Goeddel, Z. Crane, Y. Shi, Proc. Natl. Acad. Sci. USA; 2004, 101, 16, 5794–5798DOI:10.1073/pnas.0307548101
- ↑ J. L. Arbour, H. S. Rzepa, J. Contreras-García, L. A. Adrio, E. M. Barreiro, K. K. Hii, Chemistry - A European Journal, 2012, 18, 36, 11317–11324DOI:10.1002/chem.201200547
- ↑ G. A. Morris, S. B. T. Nguyen, J. T. Hupp, J. Mol. Cat. A: Chemical 174, 2001, 15–20DOI:S1381-1169(01)00165-0
- ↑ W. Zhang, J. L. Loebach, S. R. Wilson, E. N. Jacobsen, J. Am. Chem. Soc., 1990, 112, 7, 2801-2803 DOI:10.1021/ja00163a052
- ↑ M. Palucki, P. J. Pospisil, W. Zhang, E. N. Jacobsen, J. Am. Chem. SOC. 1994,116, 9333-9334DOI:10.1021/ja00099a062
- ↑ C. Altona, J. H. Ippel, A. J. A. W. Hoekzema, C. Erkelens, M. Groesbeek, L. A. Donders, A. Lambertus, Magnetic Resonance in Chemistry, 1989 , 27, 6, 564 - 576DOI:10.1002/mrc.1260270609
- ↑ 24.0 24.1 P. C. Bulman Page, B. R. Buckley ,H. Heaney, A. J. Blacker, Org. Lett., 2005, 7, 3, 375–377DOI:10.1021/ol047836h
- ↑ 25.0 25.1 25.2 L. Paquette, N. A. Pegg, D. Toops, G. D. Maynard, R. D. Rogers, J. Am. Chem. Soc., 1990, 112, 277-283 DOI:10.1021/ja00157a043
- ↑ 26.0 26.1 26.2 D. Xiong, X. Hu, S. Wang, C.-X. Miao, C. Xia, W. E. Sun, J. Org. Chem., 2011, 23, 4289–4292DOI:[4]
- ↑ 27.0 27.1 P. C. B. Page, B. R. Buckley, H. Heaney , A. J. Blacker, Org. Lett., 2005, 7, 3, 375–377 DOI:10.1021/ol047836h
- ↑ C. Guo, R. D. Shah, R. K. Dukor, X. Cao, T. B. Freedman , L. A. Nafie, Anal. Chem., 2004, 76, 23,6956–6966DOI:10.1021/ac049366a
- ↑ 29.0 29.1 29.2 29.3 29.4 29.5 29.6 G.-J.Kim, D.-W. Park, Y.-S. Tak, Catalysis Letters, 2000, 65, 1-3, 127-133DOI:10.1023/A:1019040532103
- ↑ 30.0 30.1 D. Xiong, X. Hu, S. Wang, Shoufeng, C.-X. Miao, C. Xia, W. Sun, European Journal of Organic Chemistry, 2011 , 23, 4289 - 4292
- ↑ Y. Kon, H. Hachiya, Y. Ono, T. Matsumoto, K. Sato, Synthesis, 2011, 7, 1092-1098DOI:[5]
- ↑ K. Sarma,N. Bhati, N. Borthakur, A. Goswami, Tetrahedron, 2007, 63, 36, 8735,-8741
- ↑ 33.0 33.1 S. Pedragosa-Moreau, A Archelas, R Furstoss, Tetrahedron, 1996, 52, 13, 4593–4606 DOI:10.1016/0040-4020(96)00135-4
- ↑ 34.0 34.1 34.2 34.3 M. Hayashi, H. Miwata, N. Oguni, J. Chem. Soc., Perkin Transactions 1: Organic and Bio-Organic Chemistry, 1991, 5, 1167-1171 DOI:[6]
- ↑ 35.0 35.1 V. Capriati, S. Florio, R. Luisi, A. Salomone, Org. Lett., 2002, 4, 14, 244-2448DOI:10.1021/ja9825332
- ↑ D. E. White, E. N. Jacobsen, Tetrahedron: Asymmetry, 2003, 14, 22, 3633-3638DOI:10.1016/j.tetasy.2003.09.024
- ↑ 37.0 37.1 R. Imae, N. Itoh, H. Toda, Tetrahedron Asymm., 2012, 23, 22-23, 1542-1549DOI:[7]
- ↑ P. J. Stephens, F. J. Devlin, J. R. Cheeseman, VCD Spectroscopy for Organic Chemists, CRC Press, 2012, 1-30Template:ISBN
- ↑ B. M. Bulheller, G. D. Pantoş, J. K. M. Sanders, J. D. Hirst, Phys. Chem. Chem. Phys., 2009,11, 6060-6065DOI:10.1039/B905187B
