Rep:Mod:07062
Optimisation of a molecule
Computational methods can be used to optimise the geometry of simple molecules using programmes such as Gaussian. The optimised molecule can be analysed to predict the IR absorptions and the location of the molecular orbitals.
BH3 Optimisation
B3LYP, 3-21G optimisation
A molecule of BH3 was drawn in GaussView 5.0 with a trigonal planar structure. Each of the B-H bond lengths was set to 1.5Å. The molecule was optimised with the B3LYP method and 3-21G basis set and submitted to Gaussian 09W. As the BH3 molecule is very small the calculation was fast and the level of inaccuracy would be very low. A three-dimensional image of the optimised BH3 molecule can be viewed here.

The relevant information from the LOG file is shown below. The full LOG file can be found here
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1935 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1935 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1935 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Item Value Threshold Converged? Maximum Force 0.000413 0.000450 YES RMS Force 0.000271 0.000300 YES Maximum Displacement 0.001610 0.001800 YES RMS Displacement 0.001054 0.001200 YES Predicted change in Energy=-1.071764D-06 Optimization completed. -- Stationary point found.
This shows that the optimisation was successful and complete within 4 iterations. As the Root Mean Squared (RMS) value is close to 0 this suggests the optimisation was successful and also because Gaussian has confirmed the calculation was converged. The optimised molecule had B-H bond lengths of 1.193Å and H-B-H bond angle of 120.0° which agrees well with literature.
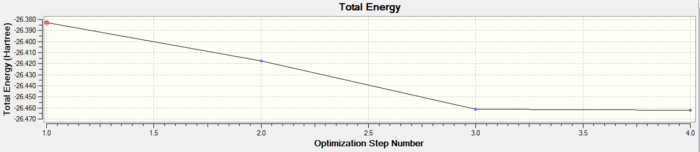
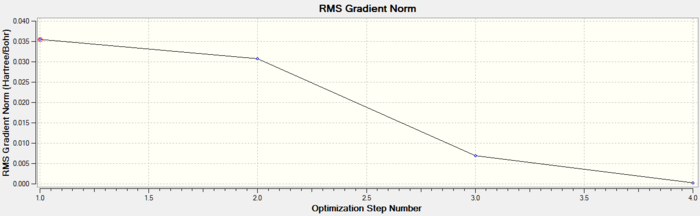
It can be seen that the RMS gradient has a very small value especially by the fourth iteration, 0.0002067 Hartree/Bohr. Once optimised the energy reaches a minimum and this further suggests that the BH3 molecule has an optimised geometry.
B3LYP, 6-31G (d,p) optimisation
The optimised BH3 molecule was oprimised again using a higher level basis set of 6-31G (d,p) instead of 3-21G which was previously used. The symmetry was turned off to ensure that the correct energy minimum was reached. The three-dimensional image of the optimised molecule can be viewed here.

The relevant information from the LOG file is shown below. The full data file can be viewed here
Item Value Threshold Converged?
Maximum Force 0.000434 0.000450 YES
RMS Force 0.000284 0.000300 YES
Maximum Displacement 0.001698 0.001800 YES
RMS Displacement 0.001114 0.001200 YES
Predicted change in Energy=-1.188111D-06
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1914 -DE/DX = 0.0004 !
! R2 R(1,3) 1.1914 -DE/DX = 0.0004 !
! R3 R(1,4) 1.1914 -DE/DX = 0.0004 !
! A1 A(2,1,3) 120.0001 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0001 -DE/DX = 0.0 !
! A3 A(3,1,4) 119.9999 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
This shows that all the displacements are converged and that the molecule has been optimised. The point group of the molecule is found to be CS which confirms that there is no symmetry. The optimised molecule had B-H bond lengths of 1.191Å and H-B-H bond angle of 120.0°.
Differences in Total Energies of Optimisation
Total energy for 3-21G optimised structure: -26.4622634 a.u.
Total energy for 6-31G (d,p) optimised structure: -26.6153226 a.u.
The energy difference between the two optimisations is 0.1530592 a.u. which seems small however when the total energy is converted to kJ/mol the energy difference is much larger between the two optimisations.
Total energy for 3-21G optimised structure: -69476.6725567 kJ/mol
Total energy for 6-31G (d,p) optimised structure: -69878.5294863 kJ/mol
There is an energy difference of -401.8569296 kJ/mol which is quite a large difference. This is because the total energy for any calculation is highly dependent on the quality of the basis set and therefore only energies for molecules with exactly the same number of atoms and with exactly the same baisis-set used on every atom can be compared.
TlBr3 Optimisation
Computational methods can be applied to TlBr3 to optimise the geometry of the molecule. TlBr3 has many more electrons than BH3 as Tl has 81 electons and Br has 35 electrons each so therefore the molecule has 186 electrons unlike BH3 which only has 8 electrons. As Tl and Br are heavier elements they both exhibit relativistic effects which cannot be recovered by the Schrodinger equation. The assumption that the valence electrons dominate in bonding interactions is used, and therefore the core electrons of an atom can be modelled by a pseudo-potential. To improve the accuracy of the optimisation we use a better basis set, LANL2DZ, which also uses pseudo-potentials.
B3LYP, LANL2DZ optimisation
A molecule a TlBr3 was drawn in GaussView 5.0 with trigonal planar geometry. The DFT-B3LYP method as used with the LANL2DZ basis set. Also the point group of the molecule was set to D3h with a very tight (0.0001) tolerance. A three-dimensional image of the optimised TlBr3 molecule can be viewed here.
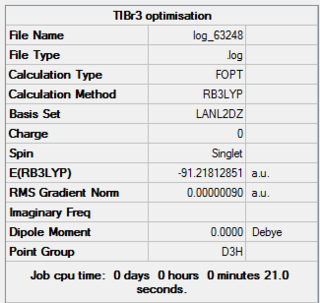
The calculation was run on A HPC server and the results from the calculation was published onto D-Space
The optimisation was successful as the calculations have all converged.
Item Value Threshold Converged? Maximum Force 0.000002 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000022 0.001800 YES RMS Displacement 0.000014 0.001200 YES Predicted change in Energy=-6.084060D-11 Optimization completed. -- Stationary point found.
The optimised molecule had Tl-Br bond lengths of 2.65095Å and Br-Tl-Br bond angle of 120.000°. The literature value reported for the length of the Tl-Br bond is 2.52Å[1] which is similar to the bond length from my calculation. However the bond length I calculated was in gas phase rather than those found in literature which are in solid or solution state. Also in the calculation the basis set did not have such a high level of accuracy so our calculated bond length may vary slightly from that found in literature.
BBr3
B3LYP, 6-31G (d,p) and LANL2DZ optimisation
BBr3 contains both heavy and light atoms and therefore a mixture of pseudo-potentials and basis sets is used.The LANL2DZ basis set was used on the bromine atoms but the 6-31G(d,p) basis set was used on the boron atom. A three-dimensional image of the optimised BBr3 molecule can be viewed here.

The relevant information from the LOG file is shown below. The full data file can be viewed here
Item Value Threshold Converged? Maximum Force 0.000008 0.000450 YES RMS Force 0.000005 0.000300 YES Maximum Displacement 0.000037 0.001800 YES RMS Displacement 0.000024 0.001200 YES Predicted change in Energy=-4.109337D-10 Optimization completed. -- Stationary point found.
The optimisation was successful as the calculations have all converged.
The optimised molecule had B-Br bond lengths of 1.93397Å and Br-B-Br bond angle of 120.002°.
Comparison of Bond Distances
| Molecule | Bond Distance |
|---|---|
| BH3 | 1.191Å |
| BBr3 | 1.934Å |
| TlBr3 | 2.651Å |
The bond distance between two atoms depends on both atoms within the bond and varies depending on the electronegativity of the atoms, the orbital overlap and the size of the atoms. The greater the orbital overlap and similarity in size of orbitals, the shorter the bond distance. Therefore the order of bond lengths between these molecules is TlBr3>BBR3>BH3 as BH3 has the smallest orbitals and greatest orbital overlap.
Changing the ligand of a molecule with the same central element alters the bond distance because there is a change in electronegativity, orbital size and orbital overlap. BH3 and BBR3 both have the same central element but differ in ligand. We can see that this has resulted in a change of bond lengths. Boron and hydrogen have a lower electronegativity difference compared to boron and bromine as orbitals have less stabilisation due to the large energy difference between the atoms. Also boron and bromine and a weaker orbital overlap than boron and hydrogen. Boron has 1s and 2p orbitals but bromine has 3d, 4s and 4p orbitals so the orbital overlap is very weak. However hydrogen only has a 1s orbital and therefore there is a much stronger orbital overlap.
Changing the central element of the molecule alters the bond distance of isostructural compounds. TlBr3 and BBR3 both have the same ligand but differ in the central element. TlBr3 has a larger bond length than BBR3. Both boron and thallium are in Group 13 of the Periodic Table sp have the same number of valence electrons and similar bonding modes. However the backbonding from bromine lone pairs to the central atom will differ depending on the central atom. Boron has smaller, less diffuse orbitals compared to thallium so has a better overlap with bromine compared to thallium and bromine. However thallium and bromine have more similar electronegativities than boron and bromine so this would give a greater stabilision. The size of thallium and bromine however is much larger than boron and bromine which means they are bonded further apart. The effect of size and orbital diffusivity have a greater effect on orbital overlap compared to electronegativity and therefore the boron and bromine bond will be stronger and therefore shorter than the thallium and boron bond.
Bonds within GaussView
A bond is a region of high electron density between two atoms with a force of attraction that holds the two atoms together. The definition of a bond is difficult to describe as it is hard to tell the extent to which atoms are bonded together. For example it is hard to tell whether a bond is single, double or triple, only by looking at the bond lengths we can figure this out.
GaussView represents bonds by the distances between the two atoms and the probability of electron density. Typically a bond is an area of high electron density and therefore bonds are shown in areas of high electron density. If GaussView does not draw a bond where expected it does not mean that no bond is present but that the distance between the two atoms is longer than what Gaussian considers to be a bond. This is common for molecules with heavier elements as they are larger and their atoms too far apart to be considered a bond.
Frequency Analysis
Frequency or vibrational analysis is the second derivative of the potential energy surface. It carried out to confirm whether the structure of the optimised molecule is a minimum. If the frequencies are all positive then thestructure is a minimum, if one of them is negative the structure is a transition state, and if any more are negative then critical point has not been found and the optimisation has failed. Also the frequency analysis provides the IR and Raman modes to compare with experiment.
BH3 vibrational analysis
The frequency calculations on the BH3 was carried out using the optimised 6-31G(d,p)file to give the structure shown here.
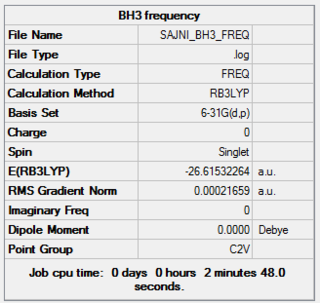
The relevant information from the LOG file is shown below.The full LOG file can be found here
Item Value Threshold Converged? Maximum Force 0.000432 0.000450 YES RMS Force 0.000217 0.000300 YES Maximum Displacement 0.001697 0.001800 YES RMS Displacement 0.000848 0.001200 YES Predicted change in Energy=-1.101958D-06 Optimization completed. -- Stationary point found.
Low frequencies --- -73.0837 -69.5094 -69.4394 -0.0007 -0.0003 0.0005
Low frequencies --- 1161.3810 1212.0964 1212.1646
1 2 3
A A A
Frequencies -- 1161.3810 1212.0964 1212.1646
Red. masses -- 1.2531 1.1072 1.1072
Frc consts -- 0.9958 0.9584 0.9585
IR Inten -- 92.7138 13.9832 13.9868
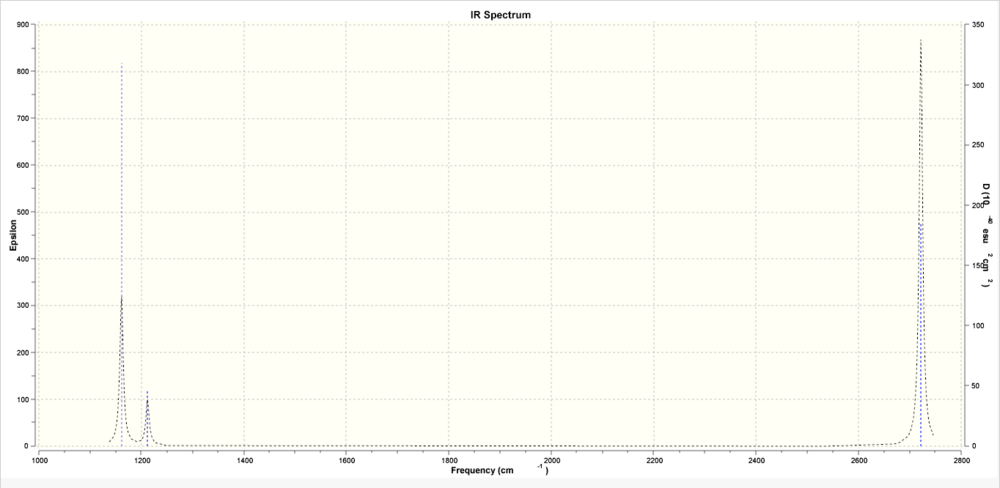
Even though there are six vibrations there are only three peaks in the spectrum because symmetric stretches are not viewed in the spectrum as vibrations must give a change in dipole for the vibration to interact with the electric field. Also the energy required for the vibrational modes of degenerate motions are equal and therefore only appear as one peak in the spectrum.
TlBr3 vibrational analysis
The frequency calculations on the TlBr3 molecule was carried out using the optimised LanL2DZ pseudo-potential file to give the structure shown here.

The calculation was run on A HPC server and the results from the calculation was published onto D-Space. The relevant information from the LOG file is shown below.
Item Value Threshold Converged? Maximum Force 0.000002 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000022 0.001800 YES RMS Displacement 0.000011 0.001200 YES Predicted change in Energy=-5.660901D-11 Optimization completed. -- Stationary point found.
Low frequencies --- -3.4213 -0.0026 -0.0004 0.0015 3.9367 3.9367
Low frequencies --- 46.4289 46.4292 52.1449
1 2 3
E' E' A2"
Frequencies -- 46.4289 46.4292 52.1449
Red. masses -- 88.4613 88.4613 117.7209
Frc consts -- 0.1124 0.1124 0.1886
IR Inten -- 3.6867 3.6867 5.8466
The lowest 'real' normal mode is a degenerate motion at 46.43 cm-1.
The optimised molecule had Tl-Br bond lengths of 2.65097Å and Br-Tl-Br bond angle of 120.000°.

Comparison of BH3 and TlBr3 Frequencies
| # | BH3 | TlBr3 |
|---|---|---|
| 1 | 1161.38 A2" | 46.43 E' |
| 2 | 1212.10 E' | 46.43 E' |
| 3 | 1212.16 E' | 52.14 A2" |
| 4 | 2587.81 A'1 | 165.27 A1' |
| 5 | 2721.51 E' | 210.69 E' |
| 6 | 2721.52 E' | 210.69 E' |
The large difference in the value of the frequencies for BH3 compared to TlBr3 indicates the large difference in the mass between the two molecules. The greater the reduced mass of a molecule the lower the vibrational frequency and also the the weaker the bond the lower the vibrational frequency. TlBr3 has a much greater reduced mass and also longer bonds, so therefore weaker bonds than BH3 which results in a lower vibrational frequency.
There has been a reordering of modes but there is still an equal number of modes because the molecules are isostructural. In both spectra the A2" and E' modes lie fairly close together, and then the A1' and E' modes also lie fairly close together but higher in energy. This is because the stretching modes are higher in energy while the bending modes are all lower in energy. In TlBr3 the A2" mode is higher in energy than the E' mode however in BH3 the A2"mode is lower in energy than the E' mode. The spectra are similar because all the other vibrational modes are the same so both spectra show a similar image of three peaks in similar positions. The peaks are slightly more defined in the BH3 spectra though.
Population Analysis
BH3
The calculation was set up in Gaussview as an energy analysis with the parameters 'pop=full'(to switch on MO analysis) and 'full nbo' selected then and run on the HPC server using the optimised 6-31G(d,p)file. The results from the calculation was published onto D-Space
BH3 Molecular Orbital Diagram
Below is the MO diagram of BH3 with a D3h point group and also the molecular orbitals calculated using Gaussian are displayed on it.

The significant differences between the real and LCAO MOs is that the real MOs has an additional MO. In LCAO methods there are only six eectrons in the molecules compared to eight electrons in the real calculation method. This results in the LUMO being one energy level greater than in the LCAO method. Also the first molecular orbital only has bonding with boron and so this means that all other molecular orbitals will be shifted an energy level higher in the real calculation method. This alters the energy levels slightly but generally the LCAO MOs and real MOs are very similar.
This suggests that that qualitative MO theory is very useful for visualising the MOs but the accuracy decreases with larger molecules as other electronic effects take place.
NBO Analysis
NH3 Optimisation
A molecule of NH3 was drawn on GaussView and optimised using the 6-31G(d,p)basis set and DFT/BYLP method.A three-dimensional image of the optimised NH3 molecule can be viewed here.
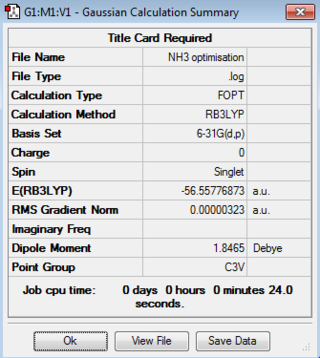
The relevant information from the LOG file is shown below. The full data file can be viewed here
Item Value Threshold Converged? Maximum Force 0.000024 0.000450 YES RMS Force 0.000012 0.000300 YES Maximum Displacement 0.000079 0.001800 YES RMS Displacement 0.000053 0.001200 YES Predicted change in Energy=-1.629727D-09 Optimization completed. -- Stationary point found.
Frequency Analysis
The frequency calculation was carried out using the optimised structure.

The relevant information from the LOG file is shown below. The full data file can be viewed here
Item Value Threshold Converged? Maximum Force 0.000021 0.000450 YES RMS Force 0.000009 0.000300 YES Maximum Displacement 0.000078 0.001800 YES RMS Displacement 0.000039 0.001200 YES Predicted change in Energy=-1.620502D-09 Optimization completed. -- Stationary point found.
Low frequencies --- -30.7788 -0.0011 -0.0008 0.0008 20.3179 28.2483 Low frequencies --- 1089.5557 1694.1235 1694.1870
The vibrational frequencies are shown below. There are no negative frequencies which ensures we have a minimum and the calculation was successful.
| Mode # | Frequency | Infrared |
|---|---|---|
| 1 | 1089.56 | 145.4390 |
| 2 | 1694.19 | 13.5560 |
| 3 | 1694.12 | 13.5557 |
| 4 | 3460.98 | 1.0593 |
| 5 | 3589.52 | 0.2709 |
| 6 | 3589.40 | 0.2700 |
Population Analysis
The calculation was set up in GaussView with the optimsied 6-31G(d,p) structure with parameters 'pop=full' and 'full nbo' slected. The calculation was then run on a HPC server and the results from the calculation was published onto D-Space
NBO Analysis

The charge limits are between -1.125 and +1.125.
Green symbolises positive while red symbolises negative.
Specific NBO charge for Nitrogen is -1.125. Specific NBO charges for Hydrogen is 0.375.
This is expected because nitrogen is highly electronegative and draws any electron density away from the hydrogen atoms.
Ammonia Borane
B3LYP, 6-31G (d,p) optimisation
A molecule of NH3BH3 was drawn on GaussView and calculation set up with a 6-31G(d,p) basis set and a DFT/B3lYP method.The three-dimensional image of the optimised molecule can be viewed here.
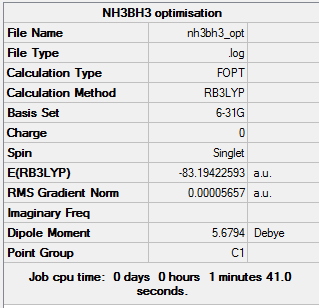
The relevant information from the LOG file is shown below. The full data file can be viewed here
Item Value Threshold Converged? Maximum Force 0.000121 0.000450 YES RMS Force 0.000057 0.000300 YES Maximum Displacement 0.000508 0.001800 YES RMS Displacement 0.000294 0.001200 YES Predicted change in Energy=-1.612062D-07 Optimization completed. -- Stationary point found.
Frequency Analysis
The frequency calculation was carried out using the optimised 6-31G(d,p) structure.The three-dimensional image of the optimised molecule can be viewed here.
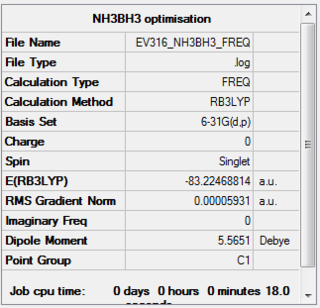
The relevant information from the LOG file is shown below. The full data file can be viewed here
Item Value Threshold Converged? Maximum Force 0.000112 0.000450 YES RMS Force 0.000059 0.000300 YES Maximum Displacement 0.000573 0.001800 YES RMS Displacement 0.000343 0.001200 YES Predicted change in Energy=-1.714294D-07 Optimization completed. -- Stationary point found.
Low frequencies --- -0.0009 -0.0001 0.0007 18.5167 23.7903 41.0221 Low frequencies --- 266.2857 632.2325 639.8311
There are no negative frequencies so the frequency calculate is said to be successul and found a minimum structure.
Energy Calculation
E(NH3)=-56.55776856 a.u. E(BH3)=-26.61532264 a.u. E(NH3BH3)=-83.22469032 a.u.
ΔE=E(NH3BH3)-[E(NH3)+E(BH3)]= (-83.2246903)-(-56.55776856+-26.61532264)= (-83.2246903)-(-83.1730912)= -0.0515991 a.u. ΔE (Dissociation Energy) = -0.0515991 a.u. = -135.4734371 kJ/mol (±10 kJ/mol)
Mini Project: Investgating Aromaticity
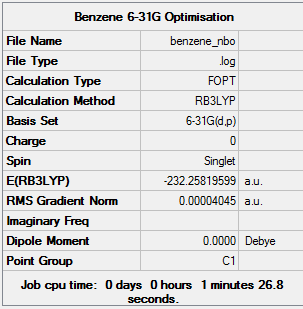
Benzene
Optimisation
The benzene molecule was first optimised using the 3-21G basis set and B3LYP method. A 3D image of the optimised structure can be viewed here. The results from the calculation was published onto D-Space
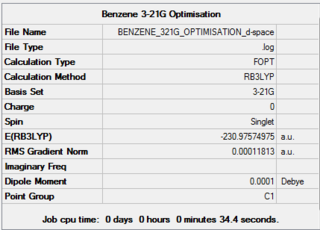
Item Value Threshold Converged? Maximum Force 0.000218 0.000450 YES RMS Force 0.000080 0.000300 YES Maximum Displacement 0.001064 0.001800 YES RMS Displacement 0.000293 0.001200 YES Predicted change in Energy=-5.021757D-07 Optimization completed. -- Stationary point found.
The benzene molecule was then optimised using the 6-31G basis set and B3LYP method. A 3D image of the optimised structure can be viewed here. The results from the calculation was published onto D-Space
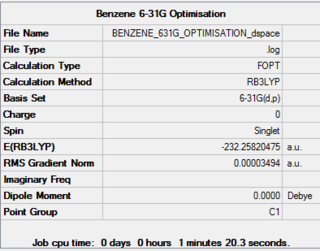
Item Value Threshold Converged? Maximum Force 0.000077 0.000450 YES RMS Force 0.000019 0.000300 YES Maximum Displacement 0.000129 0.001800 YES RMS Displacement 0.000057 0.001200 YES Predicted change in Energy=-2.461926D-08 Optimization completed. -- Stationary point found.
Frequency Analysis
The frequency calculation was carried out using the 6-31G optimised benzene molecule. A 3D image of the frequency calculated structure can be viewed here. The results from the calculation was published onto D-Space
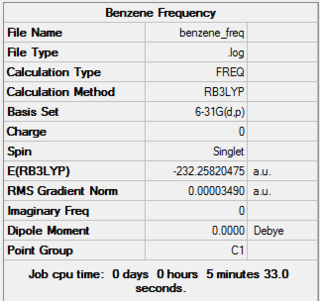
Item Value Threshold Converged? Maximum Force 0.000086 0.000450 YES RMS Force 0.000035 0.000300 YES Maximum Displacement 0.000194 0.001800 YES RMS Displacement 0.000078 0.001200 YES Predicted change in Energy=-2.794676D-08 Optimization completed. -- Stationary point found.
Low frequencies --- -12.1823 -5.8984 0.0001 0.0004 0.0010 6.5195 Low frequencies --- 414.1722 414.8763 620.9553
There are no negative frequencies so the frequency calculate is said to be successful and found a minimum structure.
NBO Analysis
The 6-31G(d,p) optimised structure was used for the calculation. The results from the calculation was published onto D-Space

The charge limits are -0.239 and 0.239.
Specific Carbon NBO charge: -0.239
Specific Hydrogen NBO charge: 0.239
MO diagram for benzene
Aromaticity and Molecular Orbitals
Huckel's Rule states that an aromatic molecule is a cyclically conjugated molecule with 4n+2 valence electrons. Benzene has 4(1)x 2 = 6 electrons and is cyclically conjugated. Only pi orbitals are cyclicaly conjugated and so only pi orbitals are aromatic. One electron from each carbon atom in benzene is delocalised into the ring.
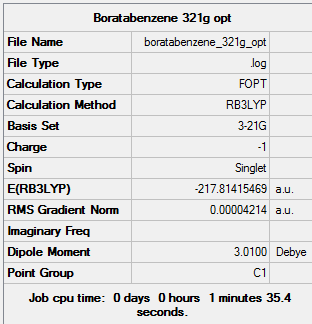
Boratabenzene
Optimisation
The boratabenzene molecule was first optimised using the 3-21G basis set and B3LYP method. A 3D image of the optimised structure can be viewed here. The results from the calculation was published onto D-Space
Item Value Threshold Converged? Maximum Force 0.000076 0.000450 YES RMS Force 0.000021 0.000300 YES Maximum Displacement 0.000286 0.001800 YES RMS Displacement 0.000083 0.001200 YES Predicted change in Energy=-4.270618D-08 Optimization completed. -- Stationary point found.
The boratabenzene molecule was then optimised using the 6-31G basis set and B3LYP method. A 3D image of the optimised structure can be viewed here. The results from the calculation was published onto D-Space

Item Value Threshold Converged? Maximum Force 0.000061 0.000450 YES RMS Force 0.000018 0.000300 YES Maximum Displacement 0.000280 0.001800 YES RMS Displacement 0.000088 0.001200 YES Predicted change in Energy=-3.751510D-08 Optimization completed. -- Stationary point found.
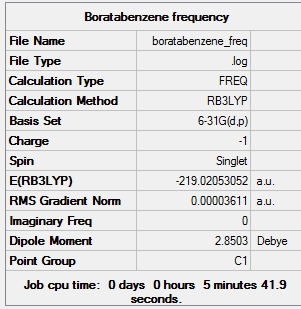
Frequency Analysis
The frequency calculation was carried out using the 6-31G optimised boratabenzene molecule. A 3D image of the frequency calculated structure can be viewed here. The results from the calculation was published onto D-Space
Item Value Threshold Converged? Maximum Force 0.000118 0.000450 YES RMS Force 0.000036 0.000300 YES Maximum Displacement 0.000319 0.001800 YES RMS Displacement 0.000129 0.001200 YES Predicted change in Energy=-4.259058D-08 Optimization completed. -- Stationary point found.
Low frequencies --- -12.4103 -0.0012 -0.0012 -0.0010 13.9747 18.0402
Low frequencies --- 371.3802 404.1485 565.1933
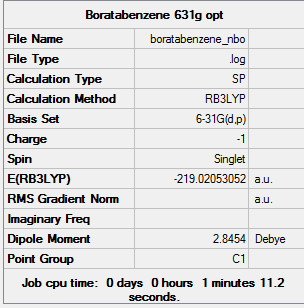
NBO Analysis
The 6-31G(d,p) optimised structure was used for the calculation. The results from the calculation was published onto D-Space
The charge limits are -0.588 and 0.588.


Pyridinium
Optimisation
The Pyridinium molecule was first optimised using the 3-21G basis set and B3LYP method. A 3D image of the optimised structure can be viewed here. The results from the calculation was published onto D-Space
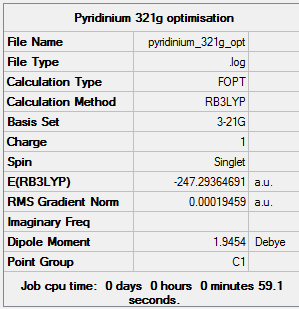
Item Value Threshold Converged? Maximum Force 0.000404 0.000450 YES RMS Force 0.000131 0.000300 YES Maximum Displacement 0.001761 0.001800 YES RMS Displacement 0.000672 0.001200 YES Predicted change in Energy=-1.648326D-06 Optimization completed. -- Stationary point found.
The pyridinium molecule was then optimised using the 6-31G basis set and B3LYP method. A 3D image of the optimised structure can be viewed here. The results from the calculation was published onto D-Space
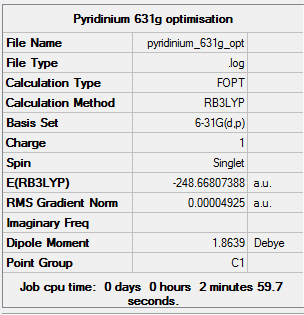
Item Value Threshold Converged? Maximum Force 0.000102 0.000450 YES RMS Force 0.000032 0.000300 YES Maximum Displacement 0.000835 0.001800 YES RMS Displacement 0.000253 0.001200 YES Predicted change in Energy=-1.441604D-07 Optimization completed. -- Stationary point found.
Frequency Analysis
The frequency calculation was carried out using the 6-31G optimised pyridinium molecule. A 3D image of the frequency calculated structure can be viewed here. The results from the calculation was published onto D-Space

Item Value Threshold Converged? Maximum Force 0.000157 0.000450 YES RMS Force 0.000049 0.000300 YES Maximum Displacement 0.000939 0.001800 YES RMS Displacement 0.000337 0.001200 YES Predicted change in Energy=-1.509366D-07 Optimization completed. -- Stationary point found.
Low frequencies --- -6.6374 -0.0012 -0.0010 -0.0004 17.2482 18.1501 Low frequencies --- 392.4864 404.0339 620.5134
NBO Analysis
The 6-31G(d,p) optimised structure was used for the calculation. The results from the calculation was published onto D-Space
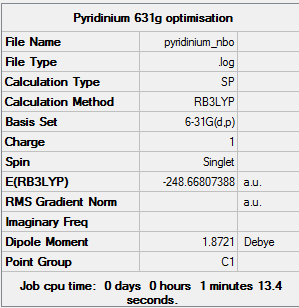
The charge limits are -0.483 and 0.483.


Borazine
Optimisation
The borazine molecule was first optimised using the 3-21G basis set and B3LYP method. A 3D image of the optimised structure can be viewed here. The results from the calculation was published onto D-Space

Item Value Threshold Converged? Maximum Force 0.000118 0.000450 YES RMS Force 0.000036 0.000300 YES Maximum Displacement 0.000351 0.001800 YES RMS Displacement 0.000101 0.001200 YES Predicted change in Energy=-1.141351D-07 Optimization completed. -- Stationary point found.
The borazine molecule was then optimised using the 6-31G basis set and B3LYP method. A 3D image of the optimised structure can be viewed here. The results from the calculation was published onto D-Space

Item Value Threshold Converged? Maximum Force 0.000124 0.000450 YES RMS Force 0.000052 0.000300 YES Maximum Displacement 0.000572 0.001800 YES RMS Displacement 0.000226 0.001200 YES Predicted change in Energy=-4.282908D-07 Optimization completed. -- Stationary point found.
Frequency Analysis
The frequency calculation was carried out using the 6-31G optimised borazine molecule. A 3D image of the frequency calculated structure can be viewed here. The results from the calculation was published onto D-Space
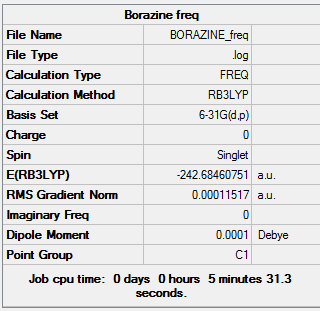
Item Value Threshold Converged? Maximum Force 0.000272 0.000450 YES RMS Force 0.000115 0.000300 YES Maximum Displacement 0.000578 0.001800 YES RMS Displacement 0.000280 0.001200 YES Predicted change in Energy=-4.304611D-07 Optimization completed. -- Stationary point found.
Low frequencies --- 0.0008 0.0011 0.0013 5.0265 11.5338 19.6156 Low frequencies --- 289.3390 289.7829 404.5807
There are no negative frequencies so the frequency calculate is said to be successful and found a minimum structure.
NBO Analysis
The 6-31G(d,p) optimised structure was used for the calculation. The results from the calculation was published onto D-Space



NBO Comparison
Benzene- there is an equal charge distribution. The carbon atoms are electronegtaive so have a negative charge while hydrogen is positive.
Boratabenzene- there is an unequal distribution of charge because of the B-H unit. B is electropositive so that the two adjacent carbon atoms have a large negative charge.
Pyridinium- there is an unequal distribution of charge because of the N-H unit. N is electronegative so that the two adjacent carbon atoms have a large positive charge. Also the two beta carbon atoms have the same charge as that on benzene.
Borazine- There is symmetry in charge distribution but the charge is not equal across all molecules. This is due to the alternating B-N bonds.
References
<references> <ref name="Tl-Br bond length">J.Blixt, J. Glase, J. Mink., J. Am. Chem. Soc., 1995, 117 (18) pp 50-89-5104.
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedTl-Br bond length