Rep:Mod:01566447
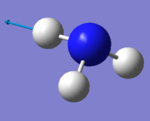
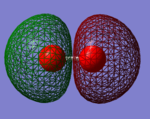
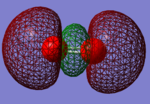
NH3 molecule
Information on optimised NH3 molecule
| Ammonia or Nitrogen trihydride | ||||
|---|---|---|---|---|
| ||||
| Calculation method: | RB3LYP | |||
| Basis set: | 6-31GP(d,p) | |||
| Final Energy: | -56.55777 au | |||
| Point group: | C3v | |||
| Optimised N-H bond length: | (1.02 ± 0.01)Å | |||
| Optimised H-N-H bond angle: | (106 ± 1)° | |||
| NBO charge on N atom | -1.125 | |||
| NBO charge on H atom | +0.375 | |||
Nitrogen is more electronegative than Hydrogen, thus it is expected to have a negative charge while the Hydrogen atoms will have positive charges.
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
The optimisation file is linked to here
NH3 Vibrations
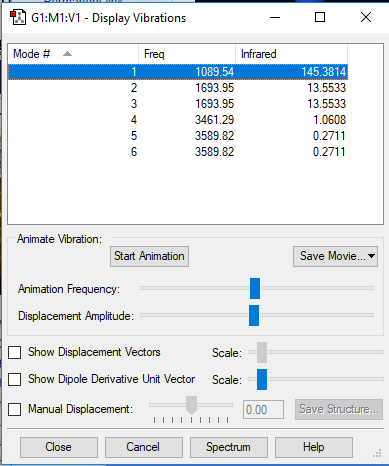
Based on the 3N-6 rule, 6 modes of vibrations are expected from an NH3 molecule. Modes with the same wavenumber are degenerate. Thus, the 2 modes of vibration corresponding to a wavenumber of 1694 cm-1, as well as the 2 modes of vibration corresponding to a wavenumber of 3590 cm-1 are degenerate. The mode of vibration with wavenumber of 1090 cm-1 is known as the "umbrella" mode. The highly symmetric mode of vibration is the one corresponding to a wavenumber of 3461 cm-1. Therefore, in an experimental spectrum of gaseous ammonia, there would be 4 bands shown on the spectrum, but only 2 bands will be visible because the other 2 bands' intensity are too low to be seen on the spectrum.
N2 molecule
Information on optimised N2 molecule
| Nitrogen molecule | ||||
|---|---|---|---|---|
| ||||
| Calculation method: | RB3LYP | |||
| Basis set: | 6-31GP(d,p) | |||
| Final Energy: | -109.52413 au | |||
| Point group: | Dh | |||
| Optimised N-N bond length: | (1.11 ± 0.01)Å | |||
| NBO charge on N atom | 0 | |||
N2 is a homodiatomic molecule, thus charge on either of the Nitrogen atoms is expected to be 0.
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
The optimisation file is linked to here
N2 Vibrations
| Wavenumber cm-1 | Symmetry | Intensity arbitrary units | Bending or bond stretch | Image |
|---|---|---|---|---|
| 2457 | SGG | 0 | Bond stretch | 
|
 | ||||
Since N2 is a linear molecule, the possible number of modes of vibration is given by 3N-5 rule, where N is the number of atoms in the molecule. 1 mode of vibration is expected from an N2 molecule. The only mode of vibration is symmetrical. No bands are expected to be seen on its spectrum because there is no change in dipole moment.
H2 molecule
Information on optimised H2 molecule
| Hydrogen molecule | ||||
|---|---|---|---|---|
| ||||
| Calculation method: | RB3LYP | |||
| Basis set: | 6-31GP(d,p) | |||
| Final Energy: | -1.17854 au | |||
| Point group: | Dh | |||
| Optimised H-H bond length: | (0.74 ± 0.01)Å | |||
| NBO charge on N atom | 0 | |||
H2 is a homo-diatomic molecule, thus charge on either of the Hydrogen atoms is expected to be 0.
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
The optimisation file is linked to here
H2 Vibrations
| Wavenumber cm-1 | Symmetry | Intensity arbitrary units | Bending or bond stretch | Image |
|---|---|---|---|---|
| 4466 | SGG | 0 | Bond stretch | 
|
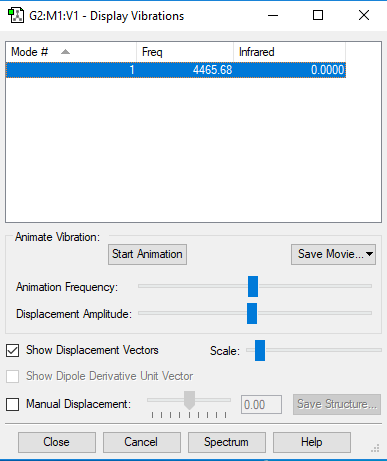 | ||||
Since H2 is a linear molecule, the possible number of modes of vibration is given by 3N-5 rule, where N is the number of atoms in the molecule. 1 mode of vibration is expected from a H2 molecule. The only mode of vibration is symmetrical. No bands are expected to be seen from its spectrum because there is no change in dipole moment.
H2 in Transition metal complexes
Using Conquest, I found Chloro-(dichloro(methyl)silyl)-bis(tricyclohexylphosphine)-(dihydrogen)-ruthenium structure, HIHVIE. The link to its structure is: https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=HIHVIE&DatabaseToSearch=Published , it has a H-H distance of 1.050Å. Then, Dihydrogen-(dihydrogenbis(3,5-bis(trifluoromethyl)pyrazolyl)-borate-H,N)-bis(tri-isopropylphosphine)-ruthenium structure, ABASUR. The link to its structure is: https://www.ccdc.cam.ac.uk/structures/Search?Ccdcid=ABASUR&DatabaseToSearch=Published , it has a H-H bond length of 1.000Å. Both are greater than the bond lengths calculated using Gaussian which is around 0.74Å, this could be due to delocalisation factors[1] in HIHVIE, or due to presence of more electron-rich ligands that induces a lengthening of the H-H bond in ABASUR.[2] Coordination with transition metals could have pulled electron density away from the Hydrogen atoms and away from the H-H bond, resulting in the weakening and lengthening of the H-H bond as well.
Haber-Bosch process
| Energy/au | |
|---|---|
| E(NH3) | -56.55777 |
| 2*E(NH3) | -113.11554 |
| E(N2) | -109.52413 |
| E(H2) | -1.17854 |
| 3*E(H2) | -3.53562 |
| ΔE=2*E(NH3)-[E(N2)+3*E(H2)] | -0.05579 |
ΔE in kJ mol-1 = 2625.5(-0.0557905) = -146.5 kJ mol-1. ΔE is energy required for converting hydrogen and nitrogen gas into ammonia gas. Ammonia gas is more stable because the chemical reaction of converting nitrogen and hydrogen into ammonia is exothermic, hence the ammonia product is lower in energy and more stable than the gaseous reactants.
CO molecule
Information on optimised CO molecule
| Carbon monoxide molecule | ||||
|---|---|---|---|---|
| ||||
| Calculation method: | RB3LYP | |||
| Basis set: | 6-31GP(d,p) | |||
| Final Energy: | -113.30945314 au | |||
| Point group: | Dh | |||
| Optimised C-O bond length: | (1.14 ± 0.01)Å | |||
| NBO charge on C atom | +0.506 | |||
| NBO charge on O atom | -0.506 | |||
Oxygen is more electronegative than carbon, thus a negative charge on O is expected whereas a positive charge is expected for C.
Item Value Threshold Converged? Maximum Force 0.000007 0.000450 YES RMS Force 0.000007 0.000300 YES Maximum Displacement 0.000003 0.001800 YES RMS Displacement 0.000004 0.001200 YES
The optimisation file is linked to here
CO Vibrations
| Wavenumber cm-1 | Symmetry | Intensity arbitrary units | Bending or bond stretch | Image |
|---|---|---|---|---|
| 2209 | SG | 68.0 | Bond stretch | 
|
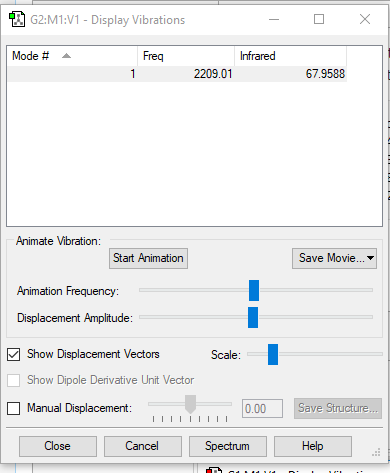 | ||||
Since CO is a linear molecule, the possible number of modes of vibration is given by 3N-5 rule, where N is the number of atoms in the molecule. 1 mode of vibration is expected from an CO molecule. The vibration is not symmetrical, there is a change in dipole moment hence 1 band is expected to be visible on its spectrum.
CO molecular orbitals
The energy between the 2s and 2p orbitals in CO is small enough to allow for sp mixing, hence the MOs in CO are a combination of the 2s and 2p orbitals of carbon and oxygen.
 [3]
[3]
O2 molecule
Information on optimised O2 molecule
| Oxygen molecule | ||||
|---|---|---|---|---|
| ||||
| Calculation method: | RB3LYP | |||
| Basis set: | 6-31GP(d,p) | |||
| Final Energy: | -150.2574243 au | |||
| Point group: | Dh | |||
| Optimised O-O bond length: | (1.22 ± 0.01)Å | |||
| NBO charge on O atom | 0 | |||
O2 is a homo-diatomic molecule, hence the charge on either of the Oxygen atoms is expected to be 0.
Item Value Threshold Converged? Maximum Force 0.000130 0.000450 YES RMS Force 0.000130 0.000300 YES Maximum Displacement 0.000080 0.001800 YES RMS Displacement 0.000113 0.001200 YES
The optimisation file is linked to here
O2 Vibrations
| Wavenumber cm-1 | Symmetry | Intensity arbitrary units | Bending or bond stretch | Image |
|---|---|---|---|---|
| 1643 | SGG | 0 | Bond stretch | 
|
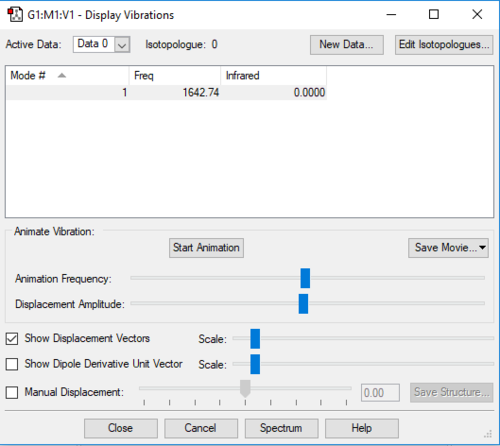 | ||||
Since O2 is a homo-diatomic linear molecule, the possible number of modes of vibration is given by 3N-5 rule, where N is the number of atoms in the molecule. 1 mode of vibration is expected from an O2 molecule. The vibration is symmetrical, there is no change in dipole moment hence no bands are expected on its spectrum.
O2 molecular orbitals
References
- ↑ S. Lachaize et al., Organometallics, 2007, 26, 3713-3721
- ↑ V. Rodriguez et al.,Organometallics 2000, 19, 2916-2926
- ↑ Chemistry Stack Exchange, https://chemistry.stackexchange.com/questions/51262/how-to-rationalise-with-mo-theory-that-co-is-a-two-electron-donor-through-carbon, (accessed March 2019)
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES - you could have discussed the energies of the MOs in more detail (number of nodes etc.)
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far