Rep:Mod:01520565
NH3 molecule
General molecular information
| Name | NH3 |
| Calculation type | FREQ |
| Calculation title | RB3LYP |
| Basis set | 6-31G(d,p) |
| E9RB3LYP) | -56.5577687 a.u. |
| RMS gradient | 0.00000485 a.u. |
| Point group | C3V |
Optimisation information
| N-H bond distance | 1.01798 |
| N-H-N bond angle | 105.741 |
Item table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986294D-10
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Jmol dynamic information
NH3 molecule |
Optimised information is linked here: File:LUVIAN NH3 OPTF POP.LOG
Vibration of NH3
Basic information about vibration
some information about vibration of NH3
According to 3N-6 rule, there should be 6 vibration modes.
Mode2 and mode3;mode5 and mode6 have the same energies, thus they are degenerate states.
Modes 1,2,3 are "bending" vibrations and modes 4,5,6 are "bond stretch" vibrations.
Modes 1 and 4 are highly symmetric.
Mode1 is known as the "umbrella" mode.
Four bands are expect to be seen in an experimental spectrum of gaseous ammonia
Specific information of each vibration mode
| Vibration mode | |
|---|---|
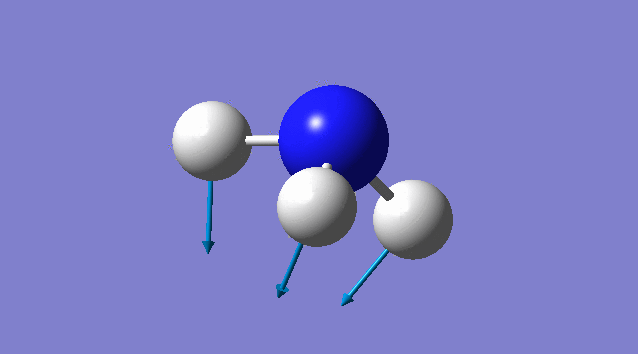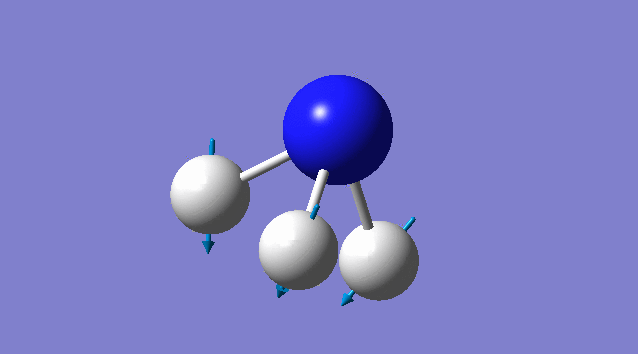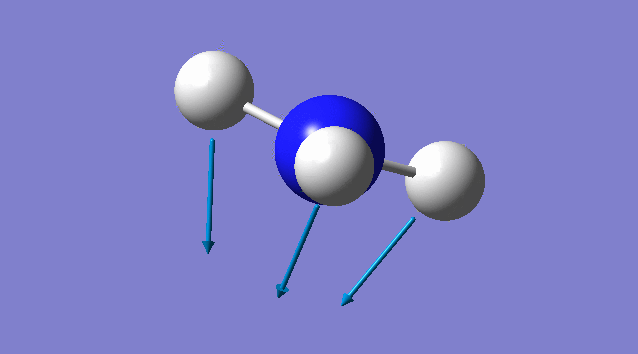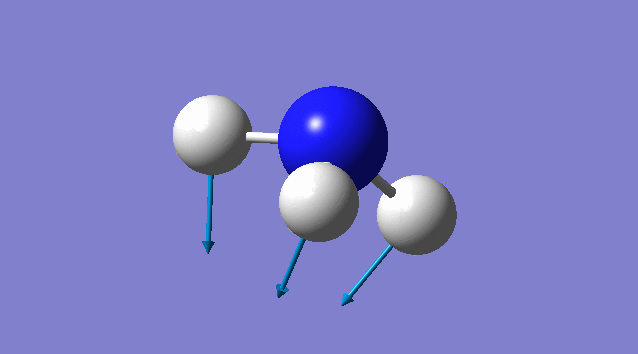
|
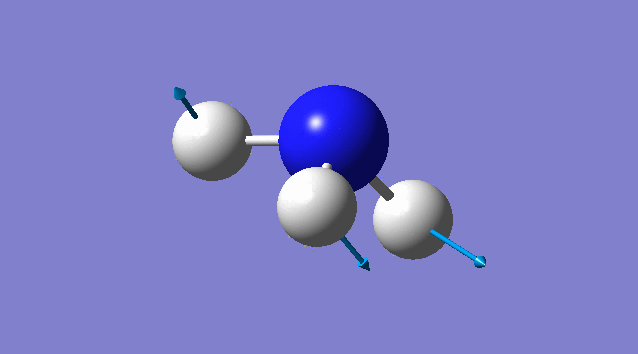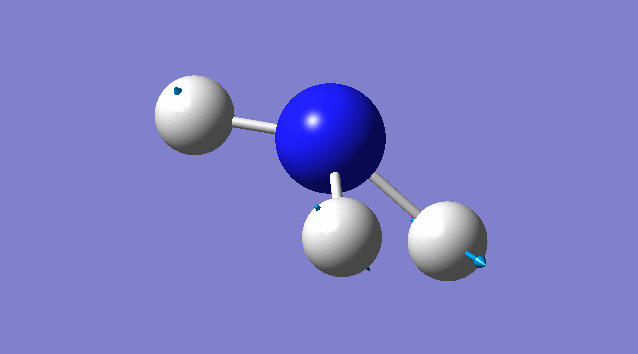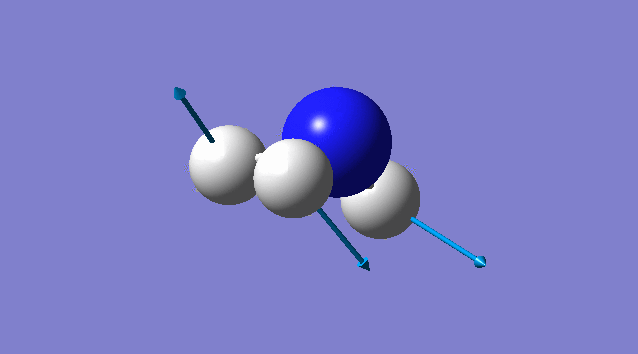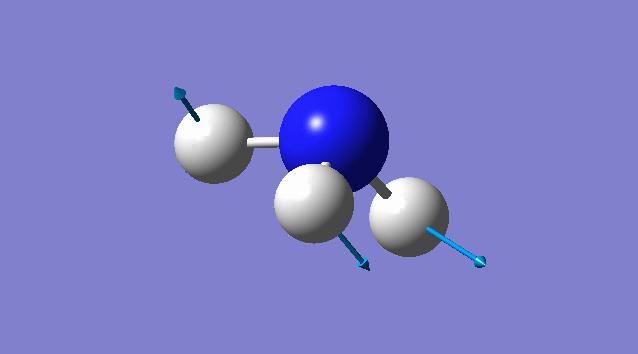
|
| wavenumber/cm-1 | |
| 1090 | 1694 |
| symmetry | |
| A1 | E |
| intensity/arbitrary units | |
| 145.4 | 13.55 |
| Vibration mode | |
|---|---|
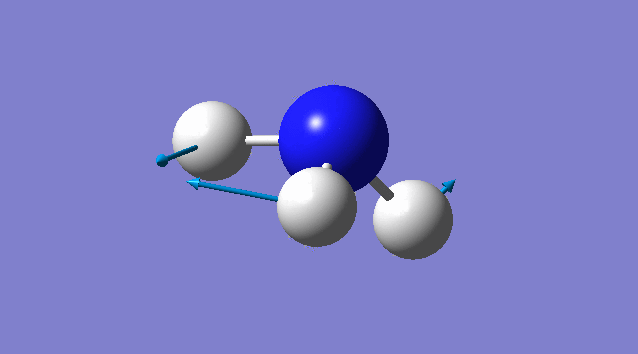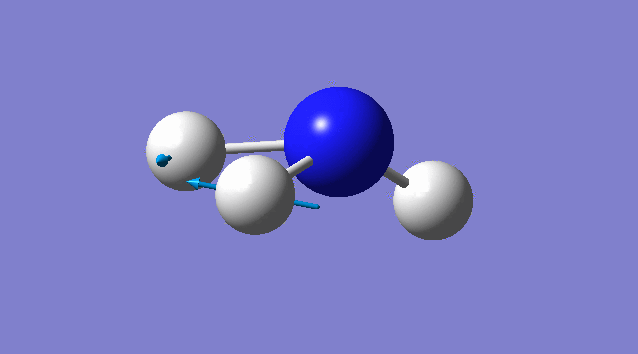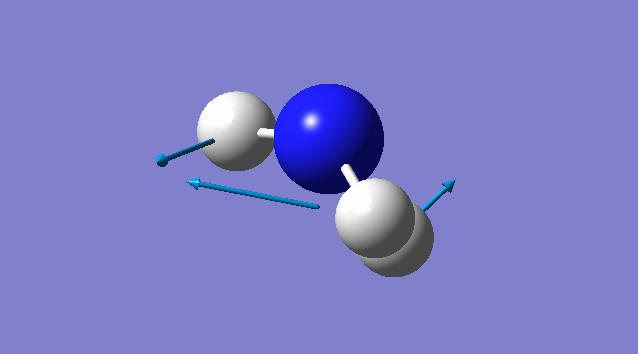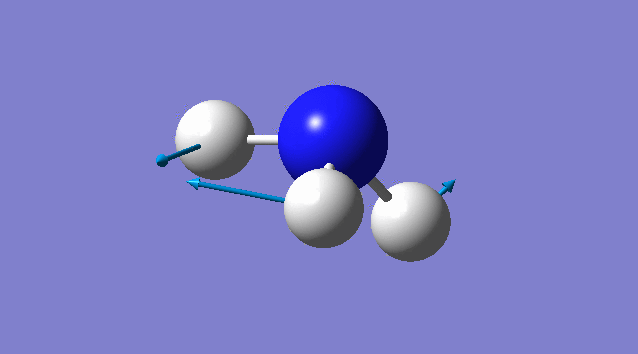
|
|
| wavenumber/cm-1 | |
| 1694 | 3461 |
| symmetry | |
| E | A1 |
| intensity/arbitrary units | |
| 13.55 | 1.061 |
| Vibration mode | |
|---|---|
|
|
| wavenumber/cm-1 | |
| 3590 | 3590 |
| symmetry | |
| E | E |
| intensity/arbitrary units | |
| 0.271 | 0.271 |
Charge distribution
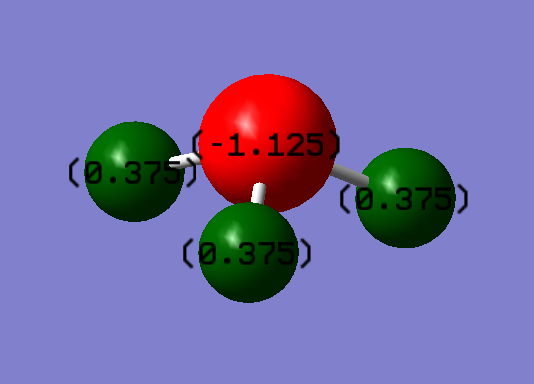
Because nitrogen has higher electronegativity than hydrogen. Thus the electron pair in the bond is pulled to the nitrogen. This results in the negative charge in N-atom and positive charge in H-atom. So the red atom is N-atom and has a -1.125 charge on it. The green atoms are H-atom and all with a charge of 0.375.
H2 molecule
General molecular information
| Name | H2 |
| Calculation type | FREQ |
| Calculation title | RB3LYP |
| Basis set | 6-31G(d,p) |
| E9RB3LYP) | -1.1785394 a.u. |
| RMS gradient | 0.0000002 a.u. |
| Point group | D*H |
Optimised information
| H-H bond distance | 0.74279 |
| bond angle | zero(linear shape) |
Item table
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Jmol dynamic information
Hydrogen molecule |
Optimised information is linked here: File:LUVIAN H2 OPT POP.LOG
Vibration of H2
Basic information about vibration
Specific information of each vibration mode
| Vibration mode |
|---|
|
| wavenumber/cm-1 |
| 4466 |
| symmetry |
| SGG |
| intensity/arbitrary units |
| 0 |
Charge distribution of H2
Because H2 is a non-polar molecule. Therefore, there is no charge on each of the hydrogen atom. Thus there will be no dipole moment.
N2 molecule
General molecular information
| Name | N2 |
| Calculation type | FREQ |
| Calculation title | RB3LYP |
| Basis set | 6-31G(d,p) |
| E9RB3LYP) | -109.5241287 a.u. |
| RMS gradient | 0.0000006 a.u. |
| Point group | D*H |
Optimised information
| N-N bond distance | 1.10550 |
| bond angle | zero(linear shape) |
Item table
Item Value Threshold Converged
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.401189D-13
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Jmol dynamic information
Nitrogen molecule |
Optimised information is linked here: File:LUVIAN N2 OPT POP.LOG
Vibration of N2
Basic information about vibration
Specific information of each vibration mode
| Vibration mode |
|---|
|
| wavenumber/cm-1 |
| 2457 |
| symmetry |
| SGG |
| intensity/arbitrary units |
| 0 |
Charge distribution of N2
Because N2 is a non-polar molecule. Therefore, there is no charge on each of the hydrogen atom. Thus there will be no dipole moment.
The mono-metallic TM complex
I found bis(dinitrogen)-(1,1,4,8,11,11-hexaphenyl-1,4,8,11-tetraphosphaundecane)-molybdenum structure AQUVOY and the two N-N distances are 1.1055 and 1.12(1) Å. The difference between computational and experimental bond lengths is because the central metal in the picked complex molecule forms a bond with the N2 legend. Thus one of the nitrogen is pulled to the metal, this increases the N-N length. Additionally, the bond length calculated by computer is the length that the molecule with a lowest energy level. But in experimental, the molecule is not always in the lowest energy state, this contributes to the difference in bond lengths. Alos, the nitrogen is in gaseous state in computational state, but the complex compound is not always in gaseous state. These reasons contribute to the difference between two bond lengths.
I find the website , here is the link https://www.ccdc.cam.ac.uk/structures/search?pid=csd:AQUVOY
Haber-Bosch process
Equation of the reaction
N2 + 3H2 -> 2NH3
Energy change of the reaction
E(NH3)=-56.5577687 a.u.
2*E(NH3)=-113.1155375 a.u.
E(N2)=-109.5241287 a.u.
E(H2)=-1.1785394 a.u.
3*E(H2)=-3.5356182 a.u.
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.0557906 a.u.=-146.5 KJ/mol
According to the calculation, the ammonia is in higher energy state than the reactants. Thus the reactants are more stable than ammonia because the energy is released. it is due to the difficulty of breaking the triple bond in nitrogen.
Cl2 molecule
General molecular information
| Name | Cl2 |
| Calculation type | FREQ |
| Calculation title | RB3LYP |
| Basis set | 6-31G(d,p) |
| E9RB3LYP) | -920.3498789 a.u. |
| RMS gradient | 0.0000251 a.u. |
| Point group | D*H |
Optimised information
| Cl-Cl bond distance | 2.04174 |
| bond angle | zero(linear shape) |
Item table
Item Value Threshold Converged?
Maximum Force 0.000043 0.000450 YES
RMS Force 0.000043 0.000300 YES
Maximum Displacement 0.000121 0.001800 YES
RMS Displacement 0.000172 0.001200 YES
Predicted change in Energy=-5.277258D-09
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.0417 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Jmol dynamic information
Clchlorine molecule |
Optimised information is linked here: File:LUVIAN CL2 OPT POP.LOG
Vibration of Cl2
Basic information about vibration
Specific information of each vibration mode
| Vibration mode |
|---|

|
| wavenumber/cm-1 |
| 520.3 |
| symmetry |
| SGG |
| intensity/arbitrary units |
| 0 |
Charge distribution of Cl2
Because Cl2 is a non-polar molecule. Therefore, there is no charge on each of the hydrogen atom. Thus there will be no dipole moment.
Molecular orbital diagrams of Cl2
The electronic configuration of a Cl-atom is: 1s22s22p63s23p5. Different molecular orbitals of Cl2 is formed from the overlap of different atomic orbitals in the two Cl-atoms.
MO of 2s bonding orbital
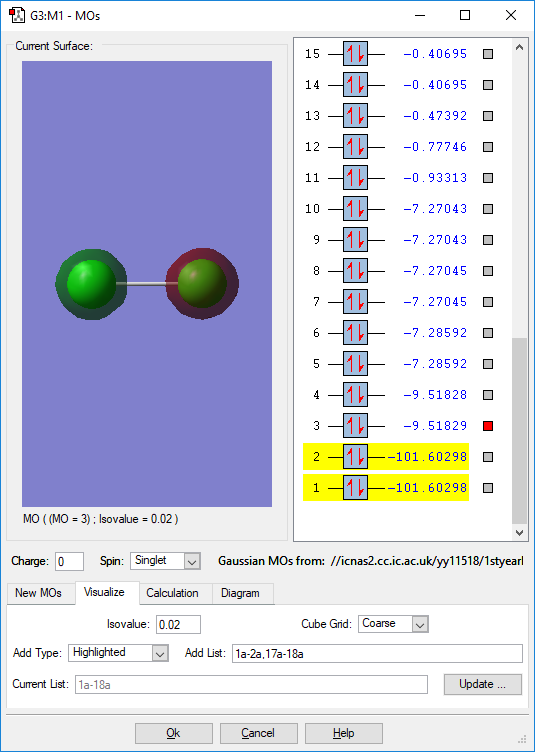
This is the picture of two atomic orbitals in each Cl-atoms. This orbital is deep in energy, which has an orbital energy of -9.051828(a.u.). Thus it is occupied by electrons. And from the picture, the orbital is close to the atom and do not have overlap. Thus do not form MO.
Because the outer lower energy electrons will first be used in the reaction. Thus, this orbital is so deep in energy and thus will not be involve in the chemical reaction.
MO of 3s bonding orbital
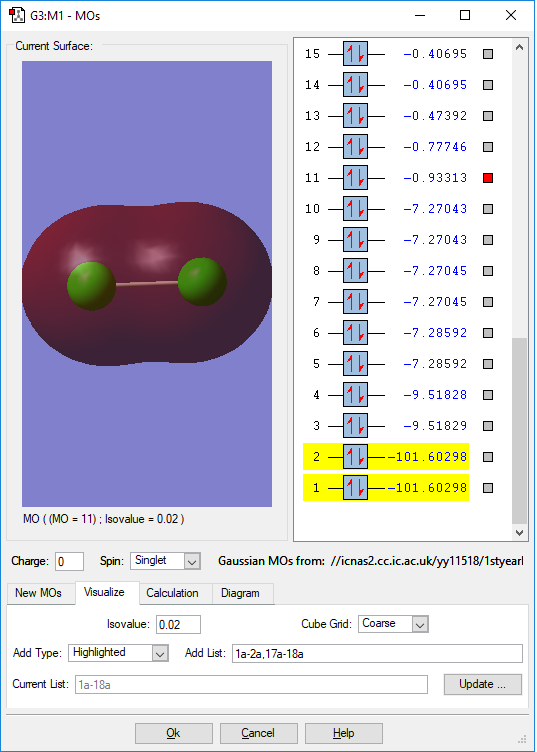
This is the 3s bonding MO of the Cl2 molecule. It is contributed from the overlap of two 3s orbitals from each chlorine atom. It has a higher energy(-0.93313 a.u.) than the upper 2s one, but it is still in lower energy level than many other orbitals and is thus be occupied. Because Cl2 molecule is SP hybridisation. This orbital is thought to be the s orbital of this hybridisation, as it is in the highest energy level among s orbitals. It will be easier to be involve in the chemical reactions.
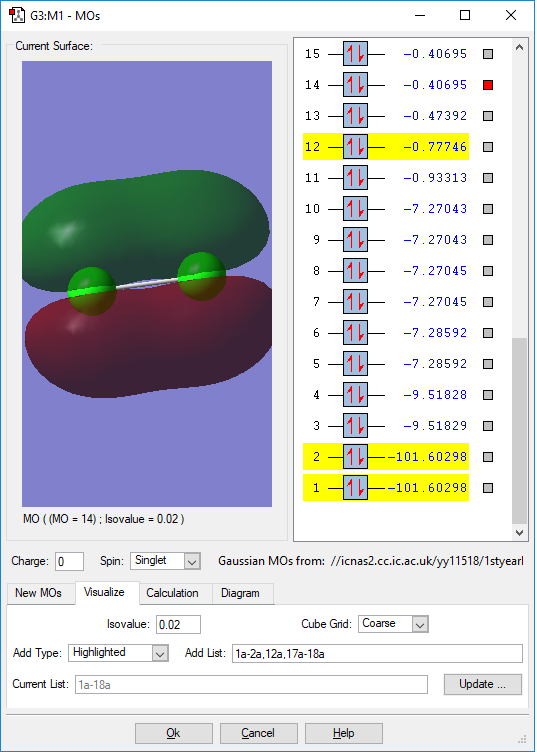
MO of 3p bonding orbital
This is the bnding orbital of 3p orbital. And this is contributed by two atomic p orbitals from each Cl atom. It has a orbital energy of -0.40695 a.u.. It is noticed that there is another MO which has the same energy with this orbital. This is because each Cl atom has two unhybridised p orbitals, the overlap between the two unhybridised p orbitals results in the two degenerate energy levels. This orbital is in higher energy level and even close to the HOMO-LUMO region. Thus it will be easily involved in the chemical reaction.
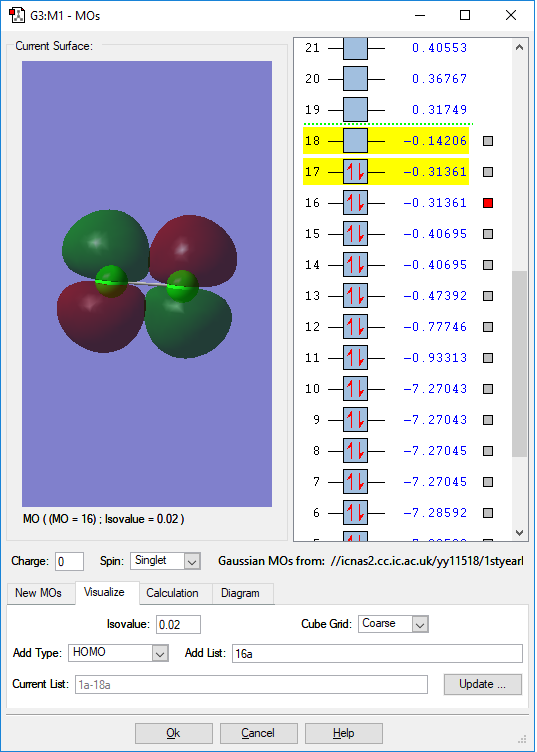
MO of HOMO
This is the HOMO(highest occupied molecular orbital) of chlorine molecule. This is the anti-bonding of the two unhybridised p orbitals in each chlorine atom, and this has a negative contribution to bonding. Because there are two unhybridised p orbitals in each chlorine atom, thus forming two anti-bondings with the same energy levels. Thus the two orbitals are both thought to be the HOMO. They both have -0.31361a.u. orbital energy. The difference between the two MOs is just the orientation.
Because they are HOMOs and has high energy, it is usually involved in the chemical reactions. Especially in the reactions having exchanges of electrons.
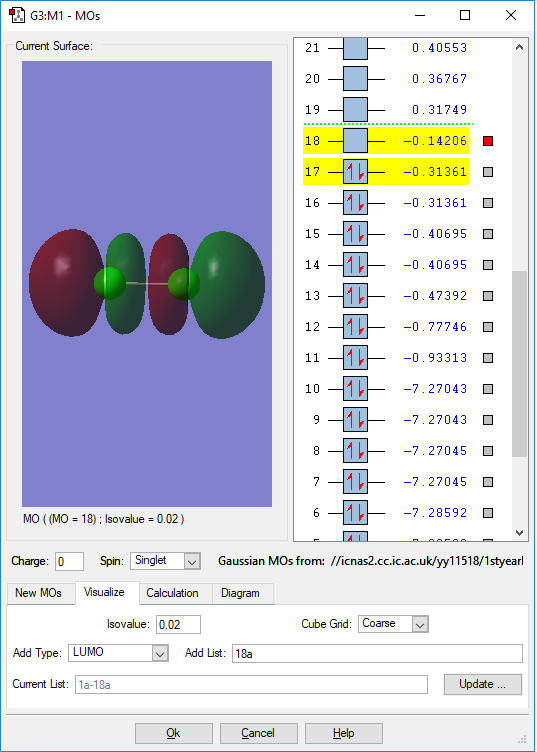
MO of LUMO
This is the LUMO(lowest unoccupied molecular orbital). This is also an anti-bonding orbital and with no electron occupied in this orbital. Because it is so high in energy(-0.14206 a.u.), it will be involved in almost all the chemical reactions. This orbital is formed when two hybridised p orbitals overlap. Because of hybridisation, the anti-bonding will be hybridised to have higher energy than other anti-orbitals formed by two unhybridised p orbitals.
BF3 molecule
General molecular information
| Name | BF3 |
| Calculation type | FREQ |
| Calculation title | RB3LYP |
| Basis set | 6-31G(d,p) |
| E9RB3LYP) | -324.5532401 a.u. |
| RMS gradient | 0.0000016 a.u. |
| Point group | D3H |
Optimisation information
| N-H bond distance | 1.31774 |
| N-H-N bond angle | 120.000 |
Item table
Item Value Threshold Converged?
Maximum Force 0.000003 0.000450 YES
RMS Force 0.000002 0.000300 YES
Maximum Displacement 0.000006 0.001800 YES
RMS Displacement 0.000004 0.001200 YES
Predicted change in Energy=-2.587589D-11
Optimization completed.
-- Stationary point found.
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.3177 -DE/DX = 0.0 !
! R2 R(1,3) 1.3177 -DE/DX = 0.0 !
! R3 R(1,4) 1.3177 -DE/DX = 0.0 !
! A1 A(2,1,3) 120.0 -DE/DX = 0.0 !
! A2 A(2,1,4) 120.0 -DE/DX = 0.0 !
! A3 A(3,1,4) 120.0 -DE/DX = 0.0 !
! D1 D(2,1,4,3) 180.0 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Jmol dynamic information
BF3 molecule |
Optimised information is linked here: File:LUVIAN BF3 OPT POP.LOG
Vibration of BF3
Basic information about vibration
Specific information of each vibration mode
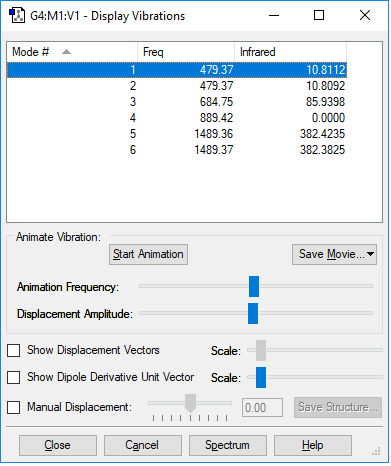
| Vibration mode | |
|---|---|

|
|
| wavenumber/cm-1 | |
| 479 | 479 |
| symmetry | |
| A | A |
| intensity/arbitrary units | |
| 10.8112 | 10.8092 |
| Vibration mode | |
|---|---|

|
|
| wavenumber/cm-1 | |
| 685 | 889 |
| symmetry | |
| A2 | B |
| intensity/arbitrary units | |
| 85.9398 | 0.0000 |
| Vibration mode | |
|---|---|
|
|
| wavenumber/cm-1 | |
| 1489 | 1489 |
| symmetry | |
| A | A |
| intensity/arbitrary units | |
| 382.4235 | 382.3825 |
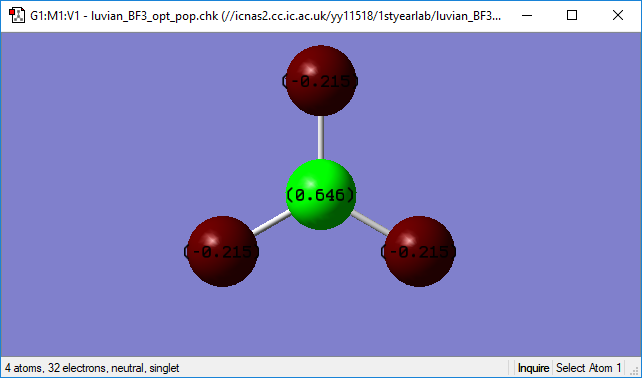
Charge distribution
Because F-atom has higher electronegativity than B-atom. Thus the electron pair in the bond is pulled to the fluorine. This results in the negative charge in F-atoms and positive charge in B-atom. So the red atoms are F-atom and all with a -0.215 charge on it. The green atoms is B-atom, with a charge of 0.646.
Marking
Note: All grades and comments are provisional and subjecct to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have recieved your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 0.5/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES, however only 2 band are expected in the experimental spectrum. You correctly stated that there are two sets of degenerate modes - this explains a spectrum with 4 peaks. However there are only 2 peaks visible as peaks 4, 5 and 6 are of too low an intensity to be visible.
N2 and H2 0/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES, you could have explained that the charges are 0 as the electronegativities are equal. You stated a bond angle for H2 and N2. To define a bond angle a minimum of 3 atoms is needed!
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4.5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
You have done a good job of presenting this information, well done! You could have explained the charges using an electronegativity argument. MO 1 is still called a MO even though there is no overlap. It is non-bonding in nature. Hybridisation is not relevant at this point to explain the bonding situation.
Independence 1/1
If you have finished everything else and have spare time in the lab you could: Check one of your results against the literature, or Do an extra calculation on another small molecule, or
YES - well done!
Do some deeper analysis on your results so far