Rep:Mod:01509348
NH3
NH3 optimisation
| molecule | NH3 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -56.55776873a.u. |
| RMS Gradient Norm | 0.00000485a.u. |
| Point Group | C3V |
| bond length of NH | 1.11 Å |
| bond angle of H-N-H | 106 degree |
bond length of NH
1.11 Å
bond angle of H-N-H
106 degree
item table
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
test molecule |
The optimisation file is liked to here
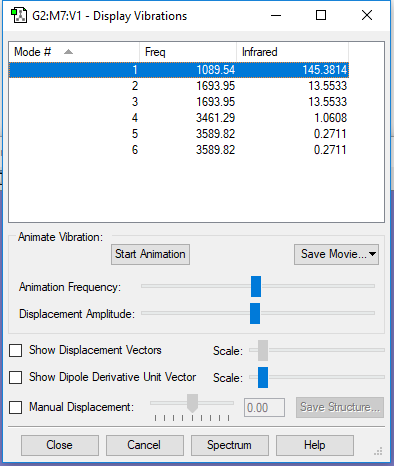
| wavenumbercm-1 | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| symmetry | A1 | E | E | A1 | E | E |
| intensity arbitrary units | 146 | 14 | 14 | 1 | 0 | 0 |
| screen shot of vibration mode |  |
 |
 |
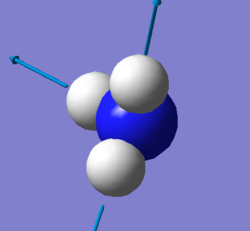 |
 |
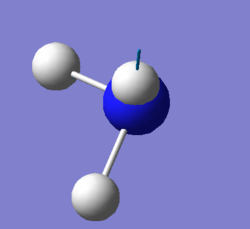
|
| number of vibration modes | 3 |
| degenerate | 1694cm-1,3590 cm-1 |
| bending | 1090cm-1,1694cm-1,1694cm-1 |
| stretching | 3461cm-1,3590cm-1,3590cm-1 |
| highly symmetric | 3461cm-1 |
| umbrella | 1090cm-1 |
| number of bands in an experimental spectrum of gaseous ammonia | 2 |
Charge of atoms in NH3
| atom | charge |
|---|---|
| N | -1.125 |
| H | 0.375 |
charge expected for N is negative as N is much more electronegative than H, so it draws majority of electron density, giving a negative charge. whereas H shows partial positive, giving a positive charge.
N2
N2 optimisation
| molecule | N2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -109.52412868a.u. |
| RMS Gradient Norm | 0.00000365a.u. |
| Point Group | D*H |
bond length of NN
1.10550 Å
item table
Item Value Threshold Converged? Maximum Force 0.000006 0.000450 YES RMS Force 0.000006 0.000300 YES Maximum Displacement 0.000002 0.001800 YES RMS Displacement 0.000003 0.001200 YES
test molecule |
The optimisation file is liked to here
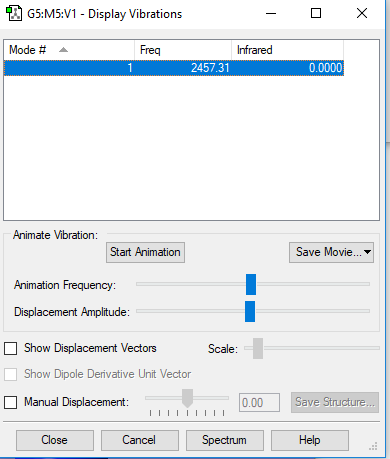
| wavenumbercm-1 | 2457 |
| symmetry | SGG |
| intensity arbitrary units | 0 |
| number of vibration mode | 1 |
| degenerate | only has one frequency of 2457 cm-1 |
| bending | N/A |
| stretching | 2457 |
| highly symmetric | 2457 |
| umbrella | N/A |
| number of bands in an experimental | N/A |
Charge of atoms in N2
| atom | charge |
|---|---|
| N | 0.000 |
| N | 0.000 |
the charge on both N atoms should be zero since there is no electronegativity in N2, no permanent dipole, even distribution of electron density
mono-metallic TM complex that coordinates N2
| unique identifier | DAYSUR |
| compound name | mer-Chloro-dinitrogen-(methylisocyanide)-tris(trimethylphosphite)-rhenium(i) |
| distance in crystal structure | 1.04 Å |
| computational distances | 1.11 Å |
The bond length crystal structure is different from computational distance. This is due to the fact that there is interaction(coordination) between the NN bond and the functional groups in the crystal structure. The dithiophosphinato ligand which is trans to N2 ligand is a better electron-donor which shortens Re-N and N-N.[[1]] [1]
H2
H2 optimisation
| molecule | H2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -1.17853936a.u. |
| RMS Gradient Norm | 0.00012170a.u. |
| Point Group | D*H |
bond length of HH
0.74 Å
Item table
Item Value Threshold Converged?
Maximum Force 0.000211 0.000450 YES
RMS Force 0.000211 0.000300 YES
Maximum Displacement 0.000278 0.001800 YES
RMS Displacement 0.000393 0.001200 YES
test molecule |
The optimisation file is liked to here
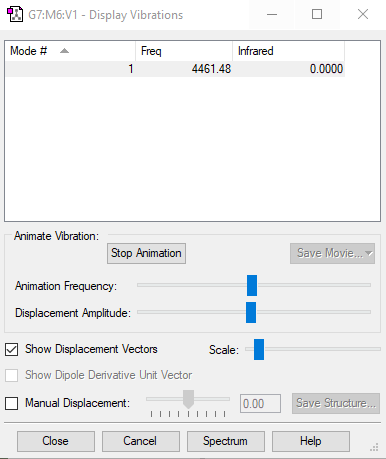
| wavenumbercm-1 | 4461 |
| symmetry | SGG |
| intensity arbitrary units | 0 |
| number of mode | 1 |
| degenerate | only has one frequency of 4461 cm-1 |
| bending | N/A |
| stretching | 4461 cm-1 |
| highly symmetric | 4461cm-1 |
| umbrella | N/A |
| number of bands in an experimental spectrum | N/A |
Charge of atoms in H2
| atom | charge |
|---|---|
| H | 0.000 |
| H | 0.000 |
the charge expected for both H2 atoms should be zero as there is no difference in electronegativity, no permanent dipole, electron density distributed evenly.
energy for Haber-Bosch process
E(NH3)=-56.55776873a.u 2*E(NH3)=-113.1155375a.u
E(N2)=-109.52412868a.u
E(H2)= -1.17853936a.u.
3*E(H2)= -3.5356179a.u
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]=-0.05579074a.u=-146.5 kJ/mol Therefore, energy for converting hydrogen and nitrogen gas into ammonia gas is -146.5 kJ/mol, product is more stable as reaction is exothermic, so product is lower in energy.
CO molecule
CO optimisation
| molecule | CO |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -113.30945314 a.u. |
| RMS Gradient Norm | 0.00000433 a.u. |
| Point Group | C*V |
| bond length of CO | 1.14 Å |
ITEM TABLE
Item Value Threshold Converged?
Maximum Force 0.000007 0.000450 YES
RMS Force 0.000007 0.000300 YES
Maximum Displacement 0.000003 0.001800 YES
RMS Displacement 0.000004 0.001200 YES
test molecule |
The optimisation file is liked to here
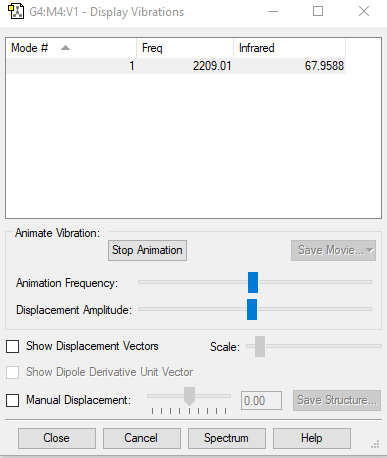
| wavenumbercm-1 | 2209 |
| symmetry | SG |
| intensity arbitrary units | 68 |
| number of vibration mode | 1 |
| degenerate | only has one frequency of 2209 cm-1 |
| bending | N/A |
| stretching | 2209 cm-1 |
| highly symmetric | 2209 cm-1 |
| umbrella | N/A |
| number of bands in an experimental spectrum | 1 |
Charge of atoms in CO
| atom | charge |
|---|---|
| C | 0.506 |
| O | -0.506 |
O is more electronegative than C, so O drags electron density to itself, showing a negative charge. C is electropositive, so it has a positive charge.
mono-metallic TM complex that coordinates CO
| unique identifier | ACOJEK |
| compound name | fac-tricarbonylchloridobis(4-hydroxypyridine)rhenium(i) pyridin-4(1H)-one solvate |
| distance in crystal structure | 1.152(5) Å,1.156(8) Å, 1.333(7) Å |
| computational distances | 1.13 Å |
Distance in crystal structure is longer than computational distance this is because of the interaction between the CO bond and rest of ligands in the crystal structure. when C binds to Re atom, it is destabilised since the electrons can not delocalised to have resonance form, causing the lengthen of CO distance.[[2]] [2]
O2
O2 optimisation
| molecule | O2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -150.25250603 a.u. |
| RMS Gradient Norm | 0.00007502 a.u. |
| Point Group | D*H |
CO2
CO2 optimisation
| molecule | CO2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -188.58093945 a.u. |
| RMS Gradient Norm | 0.00001154 a.u. |
| Point Group | D*H |
bond length of C=O in CO2
1.17Å
item table
Item Value Threshold Converged?
Maximum Force 0.000024 0.000450 YES
RMS Force 0.000017 0.000300 YES
Maximum Displacement 0.000021 0.001800 YES
RMS Displacement 0.000015 0.001200 YES
test molecule |
The optimisation file is liked to here
Charge of atoms in CO2
| atom | charge |
|---|---|
| C | 1.022 |
| O | -0.511 |
Energy of reaction involves CO
2CO+O2-> 2CO2
E(CO)=-113.30945314 a.u.
E(O2)=-150.25250603 a.u.
2*E(CO)=-226.6189063 a.u.
E(CO2)=-188.58093945 a.u.
2*E(CO2)=-377.1618789 a.u
ΔE=2*E(CO2)-(2*E(CO2)+E(O2))=-0.29046659 a.u= -762.6kJ/mol
product CO2 is more stable than reactants as the reaction is exothermic. So product CO2 is lower in energy.
molecular orbitals of CO
Reference
- ↑ M. Fernanda N. N. Carvalho, Armando J. L. Pombeiro, Ulrich Schubert, Olli Orama, Christopher J. Pickett and Raymond L. Richards J. Chem. Soc., Dalton Trans., (1985,0, 2079-2084)
- ↑ S. Argibay-Otero, R. Carballo, E.M. Vázquez-López, Acta Crystallographica Section E: Crystallographic Communications, 2017, 73, 1551, DOI: 10.1107/S2056989017013512
Independence
CH4 optimisation
| molecule | CH4 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -40.52401404 a.u. |
| RMS Gradient Norm | 0.00003263 a.u. |
| Point Group | TD |
| bond length of CH | 1.09 Å |
| bond angle of HCH | 109 degree |
item table
Item Value Threshold Converged?
Maximum Force 0.000063 0.000450 YES
RMS Force 0.000034 0.000300 YES
Maximum Displacement 0.000179 0.001800 YES
RMS Displacement 0.000095 0.001200 YES
test molecule |
The optimisation file is liked to here
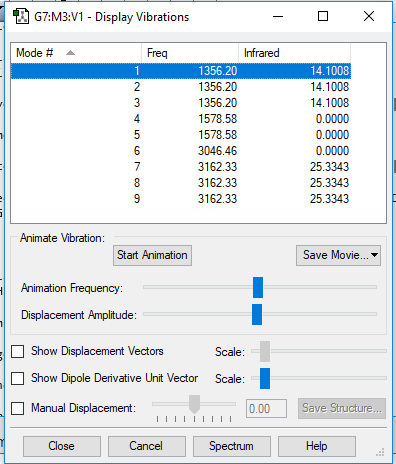
| wavenumbercm-1 | 1356 | 1356 | 1356 | 1579 | 1579 | 3046 | 3162 | 3162 | 3162 |
| symmetry | T2 | T2 | T2 | E | E | A1 | T2 | T2 | T2 |
| intensity arbitrary units | 14 | 14 | 14 | 0 | 0 | 0 | 25 | 25 | 25 |
Charge of atoms in CH4
| atom | charge |
|---|---|
| C | -0.470 |
| H | 0.118 |
H2O
H2O optimisation
| molecule | H2O |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| E(RB3LYP) | -76.41973740 a.u. |
| RMS Gradient Norm | 0.00006276 a.u. |
| Point Group | C2V |
Energy of reaction involves CH4
CH4+H2O-> CO+3H2
E(CH4)=-40.52401404 a.u.
E(H2O)=-76.41973740 a.u.
E(CO)=-113.30945314 a.u.
E(H2)=-1.17853936a.u.
3*E(H2)=-3.53561808a.u ΔE=(E(CO)+3*E(H2))-(E(CH4)+E(H2O))=0.09868022a.u.=259.1kJ/mol
the reactants are more stable than products. reaction is endothermic, reactants are lower in energy.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 0.5/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES, overall very good, however you have left all the jmol captions as the default “test molecule” this gives the reader no information.
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 5/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES, excellent explanations on the MOs, well done!
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
Do some deeper analysis on your results so far
You did extra calculations, and an extra energy comparison, and an extra literature comparison, well done!