Rep:Mod:01508310
NH3 Structural Information
N-H bond length: 1.02 Å
H-N-H bond angle: 106°
Structure: trigonal pyramidal
NH3 |
Gaussian Optimisation
Calculation method: RB3LYP
Basis set: 6-31H(d.p)
Final energy E in atomic units: -56.55776873 au
The point group of NH3: C3V
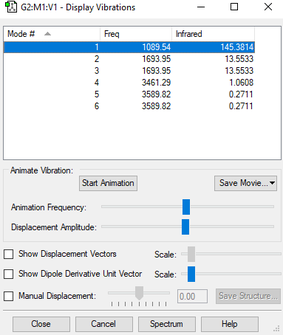
Display Vibrations
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000072 0.001800 YES RMS Displacement 0.000035 0.001200 YES
The optimisation file is linked to File:HHE NH3 OPTF-POP.LOG
Wavenumber, Symmetry and Intensity of Vibrations
Number of modes using 3N-6: 3×4-6=6
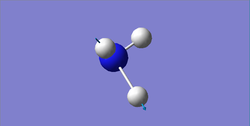
Degenerate modes: 1694 cm-1, 3590 cm-1
Bending: 1090, 1694 cm-1
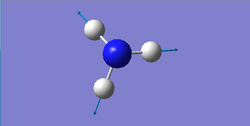
Stretching: 3461, 3590 cm-1
Highly symmetric: 3461 cm-1
Umbrella mode: 1090 cm-1
IR bands: 2 bands expected as two of them are degenerate and 3 of them are of 0 or negligible intensity.
| wavenumber (cm-1) | 1090 | 1694 | 1694 | 3461 | 3590 | 3590 |
| symmetry | A1 | E | E | A1 | E | E |
| intensity (arbitrary units) | 145 | 14 | 14 | 1 | 0 | 0 |
| image | 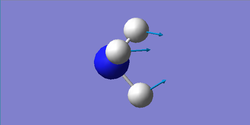 |
 |
 |
 |
 |
|
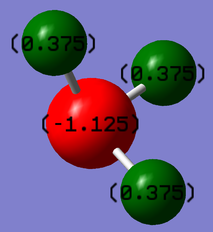
Charge Analysis
N is expected to be negative and H is expected to be positive because N is more electronegative than H and pulls electrons more towards N itself.
N2 Structural Information
Bond length: 1.11 Å
Structure: linear
N2 |
Gaussian Optimisation
Calculation method: RB3LYP
Basis set: 6-31G(d.p)
Final energy E in atomic units: -109.52412868 au
The point group of NH3: D∞h
Display Vibrations
Item Value Threshold Converged? Maximum Force 0.000145 0.000450 YES RMS Force 0.000145 0.000300 YES Maximum Displacement 0.000045 0.001800 YES RMS Displacement 0.000064 0.001200 YES
The optimisation file is linked to File:HHE N2 OPTF POP.LOG
Wavenumber, Symmetry and Intensity of Vibrations
Number of modes using 3N-5: 3×2-5=1
Degenerate modes: 0
Bending: 0
Stretching: 1
IR bands:0 because there is no change in dipole moment of the molecule in the stretching mode
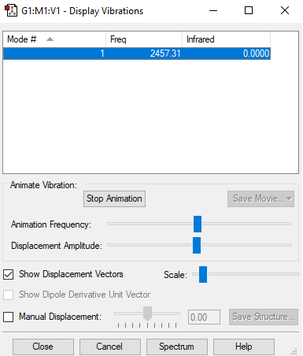
| wavenumber (cm-1) | 2457 |
| symmetry | SGG |
| intensity (arbitrary units) | 0 |
| image | 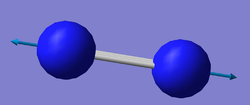
|
Charge Analysis
The charges are expected to be zero because there is no difference in electronegativities between the 2 atoms.
H2 Structural Information
Bond length: 0.74 Å
Structure: linear
H2 |
Gaussian Optimisation
Calculation method: RB3LYP
Basis set: 6-31G(d.p)
Final energy E in atomic units: -1.17853930 au
Point group: D∞h
Display vibrations
Item Value Threshold Converged? Maximum Force 0.000211 0.000450 YES RMS Force 0.000211 0.000300 YES Maximum Displacement 0.000278 0.001800 YES RMS Displacement 0.000393 0.001200 YES
The optimisation file is linked to File:HHE H2 OPTF POP.LOG
Wavenumber, Symmetry and Intensity of Vibrations
Number of modes using 3N-5: 3×2-5=1
Degenerate modes: 0
Bending: 0
Stretching: 1
IR bands: 0 because there is no change in dipole moment of the molecule in the stretching mode
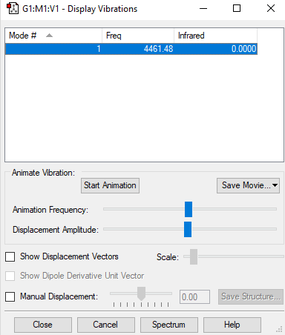
| wavenumber (cm-1) | 4461 |
| symmetry | SGG |
| intensity (arbitrary units) | 0 |
| image | 
|
Charge Analysis

The charges are expected to be zero because there is no difference in electronegativities between the 2 atoms.
Transition Metal Complex
Identifier: JISZOA[1]
Calculated bond length: 0.74 Å
Bond length in complex: 1.03 Å
In this complex, H2 donates electrons to the central metal atom and the bond energy of H2 is lower and therefore the bond length is longer.[1]
Haber-Bosch Process
E(NH3)= -56.5577687 au
2*E(NH3)= -113.1155374 au
E(N2)= -109.5241287 au
E(H2)= -1.1785393 au
3*E(H2)= -3.5356179 au
ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.0557909 au
= -146.8 kJmol-1
The ammonia product is more stable because the reaction is exothermic.
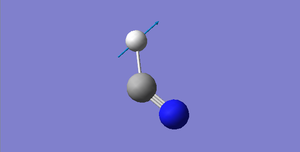
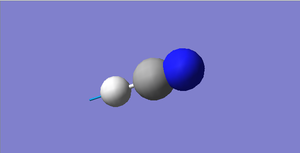
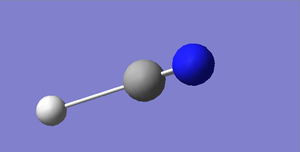
HCN Structural Information
H-N bond length: 1.07 Å
C≡N bond length: 1.16 Å
H-C≡N bond angle: 180 °
HCN |
Gaussian Optimisation
Calculation method: RB3LYP
Basis set: 6-31G(d.p)
Final energy E in atomic units: -93.42458151 au
The point group of NH3: C∞v
Display vibrations
Item Value Threshold Converged?
Maximum Force 0.000179 0.000450 YES
RMS Force 0.000094 0.000300 YES
Maximum Displacement 0.000145 0.001800 YES
RMS Displacement 0.000101 0.001200 YES
The optimisation file is linked to File:HHE HCN OPTF POP.LOG
Wavenumber, Symmetry and Intensity of Vibrations
Number of modes using 3N-5: 3×3-5=4
Degenerate modes: 769 cm-1
Bending: 769 cm-1
Stretching: 2214 and 3476 cm-1
IR bands: 3 because all vibrations cause a change in dipole moment and 2 of them are degenerate.
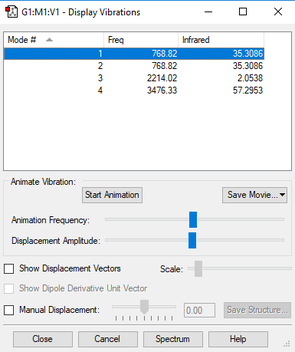
| wavenumber (cm-1) | 769 | 769 | 2214 | 3476 |
| symmetry | PI | PI | SG | SG |
| intensity (arbitrary units) | 35 | 35 | 2 | 57 |
| image |  |
 |
 |
|
Charge Analysis
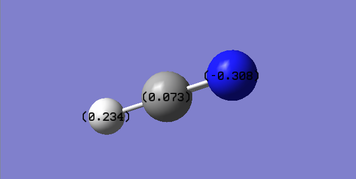
Nitrogen is the most electronegative atom of all three atoms and it is expected to have a negative charge. Carbon is more electronegative than hydrogen and therefore the charge on carbon is less positive than that on hydrogen.
Molecular Orbitals
Orbital 1
Energy:-14.36052 au
This orbital is the core 1s orbital on N which is too deep in energy to be involved in bonding. Therefore it does not contribute to the bonding in the molecule. It is occupied by 2 electrons.
Orbital 2
Energy:-10.24613 au
This is the core 1s orbital on C which is also too deep in energy to be involved in bonding. This atomic orbital does not contribute to the bonding in the molecule either. This orbital is also occupied by 2 electrons.
Orbital 3
Energy: -0.91992 au
This is a bonding molecular orbital composed of 2s atomic orbitals on N and C and a 1s atomic orbital on H. It is involved in bonding as its energy is not as deep in energy as the last 2 orbitals. All atomic orbitals are in phase and the electron density therefore encompasses the whole molecule with no nodal plane. It is occupied by 2 electrons.
Orbital 7
Energy: -0.35936 au
This is the bonding molecular orbital made up of 2py atomic orbitals on N and C. The 2 orbitals combine in phase and produce a bonding pi orbital. The hydrogen 1s atomic orbital is not involved as there is a nodal plane across the bond. It is also the HOMO, which is filled with 2 electrons.
Orbital 8
Energy: 0.01927 au
This is the LUMO and it is an antibonding orbital. It is formed by the 2px atomic orbitals on N and C, the hydrogen 1s atomic orbital is not involved for the same reason stated above. This molecular antibonding orbital is orthogonal to the one above as the px and py are orthogonal to each other. It is positioned this way just to be seen more clearly. It is empty and therefore does not contribute to the bonding in the molecule.
Independent Work
CH4 Structural Information
C-H bond length: 1.09 Å
H-C-H bond angle: 109°
Structure: tetrahedral
CH4 |
Gaussian Optimisation
Calculation method: RB3LYP
Basis set: 6-31H(d.p)
Final energy E in atomic units: -40.52401404 au
The point group of CH4: Td
Display Vibrations
Item Value Threshold Converged?
Maximum Force 0.000063 0.000450 YES
RMS Force 0.000034 0.000300 YES
Maximum Displacement 0.000179 0.001800 YES
RMS Displacement 0.000095 0.001200 YES
The optimisation file is linked to File:HHE CH4 OPTF.LOG
Wavenumber, Symmetry and Intensity of Vibrations
Number of modes using 3N-6: 3×5-6=9
Degenerate modes: 1356 cm-1, 1579 cm-1, 3162 cm-1
Bending: 1356 cm-1, 1579 cm-1
Stretching: 3046cm-1, 3162 cm-1
IR bands: 2 as 5 of them are degenerate and 3 of them are of 0 intensity.
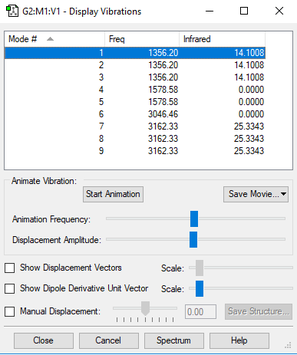
| wavenumber (cm-1) | 1356 | 1356 | 1356 | 3046 | 1579 | 1579 | 3162 | 3161 | 3162 |
| symmetry | T2 | T2 | T2 | A1 | E | E | T2 | T2 | T2 |
| intensity (arbitrary units) | 14 | 14 | 14 | 0 | 0 | 0 | 25 | 25 | 25 |
| image |  |
 |
 |
 |
 |
 |
 |
 |
|
Charge Analysis
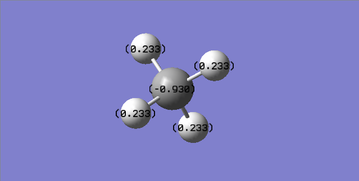
C is expected to be negative and H is expected to be positive because C is more electronegative than H and pulls electrons more towards C itself.
Reference
- ↑ Chaudret, B., Chung, G., Eisenstein, O., Jackson, S., Lahoz, F. and Lopez, J. (1991). Journal of the American Chemical Society, 113(6), pp.2314-2316.
Marking
Note: All grades and comments are provisional and subject to change until your grades are officially returned via blackboard. Please do not contact anyone about anything to do with the marking of this lab until you have received your grade from blackboard.
Wiki structure and presentation 1/1
Is your wiki page clear and easy to follow, with consistent formatting?
YES
Do you effectively use tables, figures and subheadings to communicate your work?
YES
NH3 1/1
Have you completed the calculation and given a link to the file?
YES
Have you included summary and item tables in your wiki?
YES
Have you included a 3d jmol file or an image of the finished structure?
YES
Have you included the bond lengths and angles asked for?
YES
Have you included the “display vibrations” table?
YES
Have you added a table to your wiki listing the wavenumber and intensity of each vibration?
YES
Did you do the optional extra of adding images of the vibrations?
YES
Have you included answers to the questions about vibrations and charges in the lab script?
YES
N2 and H2 0.5/0.5
Have you completed the calculations and included all relevant information? (summary, item table, structural information, jmol image, vibrations and charges)
YES
Crystal structure comparison 0.5/0.5
Have you included a link to a structure from the CCDC that includes a coordinated N2 or H2 molecule?
YES
Have you compared your optimised bond distance to the crystal structure bond distance?
YES
Haber-Bosch reaction energy calculation 1/1
Have you correctly calculated the energies asked for? ΔE=2*E(NH3)-[E(N2)+3*E(H2)]
YES
Have you reported your answers to the correct number of decimal places?
YES
Do your energies have the correct +/- sign?
YES
Have you answered the question, Identify which is more stable the gaseous reactants or the ammonia product?
YES
Your choice of small molecule 4/5
Have you completed the calculation and included all relevant information?
YES
Have you added information about MOs and charges on atoms?
YES
The information you have given I this sections are correct! MO 1 and 2 could have been labelled as non-bonding but you explained the influence on bonding correctly. You could have explained the relative MO energies (e.g. comparing number of nodes, electronegativities etc.)
Independence 1/1
If you have finished everything else and have spare time in the lab you could:
Check one of your results against the literature, or
Do an extra calculation on another small molecule, or
YES
Do some deeper analysis on your results so far





