Rep:Mod:01391805
NH3 optimisation
test molecule |
A molecule of NH3 was drawn and optimised using GaussView 5.0.9. The method of optimisation was B3LYP, the basis set was 6-31G(d,p) and the symmetry of the NH3 was set to C3v. The final energy of the molecule was -56.56au and the RMS gradient was 0.00000485au. The bond length was determined to be 1.01798Å while the dipole moment was 1.8466D and the bond angle was 105.7°.
Item Table
Item Value Threshold Converged? Maximum Force 0.000004 0.000450 YES RMS Force 0.000004 0.000300 YES Maximum Displacement 0.000070 0.001800 YES RMS Displacement 0.000033 0.001200 YES
The optimisation file is liked to here
Vibrations
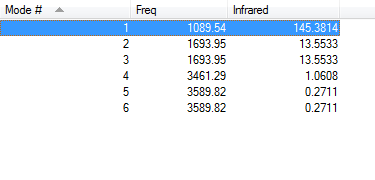
The number of vibrational modes displayed here follows the 3N-6 rule as NH3 is expected to have 6 vibrational modes. Of the 6 modes 2 and 3 are degenerate to eachother while 5 and 6 are also degenerate to eachother. The 3 lower energy modes (modes 1,2 and 3) are bending vibrations while the 3 higher energy modes (4,5 and 6) are stretching vibrations. Vibration mode 4 is highly symmetric and mode 1 is the "umbrella" mode. In an experimental spectrum of gaseous NH3 only 2 bands would be expected as the infrared integration peaks of 1.09608 for mode 4 and 0.2711 for modes 5 and 6 are too small to appear in an experimental IR spectrum where the baseline wouldn't be exactly 0.
The charge of the N atom in NH3 was found to be -1.125 while the charge on the 3 H atoms was found to be 0.375. This follows my expectations of the N atom to be negatively charged while the H atoms are positively charged as nitrogen is more electronegative than hydrogen so there would be a dipole in the N-H bond.
N2 & H2
A molecule of N2 and a molecule of H2 was optimised using GaussView under indentical settings. The method used was B3LYP, the basis set was the basis set was 6-31G(d,p) and the symmetry of both molecules was set to D∞h.
N2 optimisation
test molecule |
The optimisation file is liked to here
The final energy of the N2 molecule was found to be -109.5au and the RMS gradient was 0.00000060au. The bond length was found to be 1.10550Å and the dipole moment was 0. The actual bond length of the N2 molecule is 1.0975 ± 0.0001Å(1) which is slightly lower than what was determined using Gaussview.
N2 item table
Item Value Threshold Converged? Maximum Force 0.000001 0.000450 YES RMS Force 0.000001 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000000 0.001200 YES
This table shows the structure successfully converged.
N2 vibrations
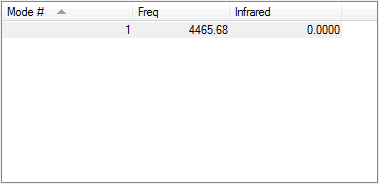
There is only 1 vibrational mode in N2, it is a symmetrical stretching vibration so would not appear on an IR spectrum.
H2 optimisation
test molecule |
The optimisation file is liked to here
The final energy of the H2 molecule was found to be -1.179au and the RMS gradient was 0.00000017au. The bond length was found to be 0.74279 with a dipole moment of 0.
H2 item table
Item Value Threshold Converged? Maximum Force 0.000000 0.000450 YES RMS Force 0.000000 0.000300 YES Maximum Displacement 0.000000 0.001800 YES RMS Displacement 0.000001 0.001200 YES
This table shows the structure successfully converged.
H2 vibrations
There is only 1 vibrational mode in H2, it's symmetrical stretching vibration so doesn't appear on its IR spectrum.
Determining the energy of the Haber-Bosch reaction
E(NH3) = -56.55776873au 2*E(NH3) = -113.11553746au E(N2) = -109.52412868au E(H2) = -1.17853936au 3*E(H2) = -3.53561808au ΔE = 2*E(NH3)-[E(N2)+3*E(H2)]= -0.05579070au ΔE = 2625.5*-0.05579069999998865 = -146.48kJ/mol
The energy for converting hydrogen and nitrogen gas into ammonia gas is -146.48kJ/mol so the ammonia is more stable than the gaseous reactants as the ammonia lies lower in energy.
ClF optimization
A molecule of ClF was optimized using GaussView using B3LYP as the method and 6-31G(d,p) as the basis set. The symmetry of the ClF molecule was set to C∞v.
test molecule |
The optimisation file is liked to here
The final energy of the ClF was found to be -559.9au and the RMS gradient is 0.00014211au. The charge on the F atom is -0.309 while the charge on the Cl atom is 0.309, the resulting dipole moment was found to be 0.9787D while the bond length was determined to be 1.66434Å.
ClF item table
Item Value Threshold Converged? Maximum Force 0.000246 0.000450 YES RMS Force 0.000246 0.000300 YES Maximum Displacement 0.000433 0.001800 YES RMS Displacement 0.000612 0.001200 YES
This item table shows that the molecule successfully converged.
ClF Vibrations
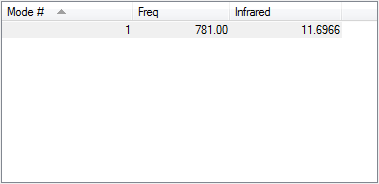
ClF only has 1 stretching vibration which is symmetrical, however the molecule already has a dipole so by stretching the bond the dipole moment changes, thus this stretch appears in the IR spectrum.
ClF Molecular Orbitals
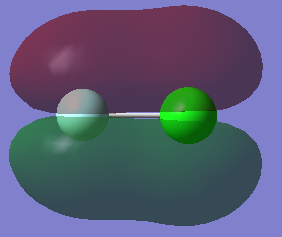
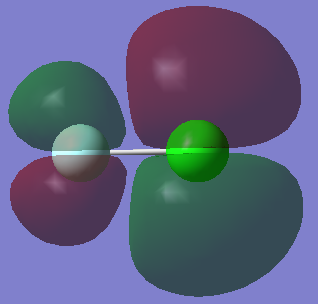
In these generated models of ClF molecular orbitals the light blue sphere represents the F atom while the green sphere represents the Cl atom.
This is a fully occupied bonding orbital, it's the result of a sigma overlap between the 2s atomic orbital of the F atom and the 3s atomic orbital of the Cl atom.
This is a fully occupied bonding orbital which is the result of a sigma overlap between the 2p atomic orbital on the F atom and a 3p atomic orbital of the Cl atom.
This is a fully occupied bonding orbital which is the result of a pi overlap between the 2p atomic orbital on the F atom and a 3p atomic orbital of the Cl atom.
This is the highest energy occupied molecular orbital (HOMO). It is an antibonding orbital which is the result of a pi overlap between a 2p atomic orbital of F and a 3p atomic orbital of Cl. The HOMO being an antibonding orbital indicates that the bond is easily broken and therefore ClF would be very reactive.
This is the lowest energy unoccupied molecular orbital. It's an antibonding orbital which is the result of a sigma overlap between the 2p atomic orbital of F and the 3p atomic orbital of Cl.
References
1. Tables of Interatomic Distances and Configuration in Molecules and Ions, L.E. Sutton, ed., London: The Chemical Society, 1958