Rep:Mod:01386123
NH3 Molecule
Molecule Name : Ammonia
Calculation Method : RB3LYP
Basis Set : 6-31G(d,p)
Final Energy E(RB3LYP) : -56.55776873 a.u.
RMS Gradient : 0.00000485 a.u.
Point Group : C3V
N-H Bond Distance : 1.01798
H-N-H Bond Angle : 105.741
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
NH molecule |
The optimisation file is linked here
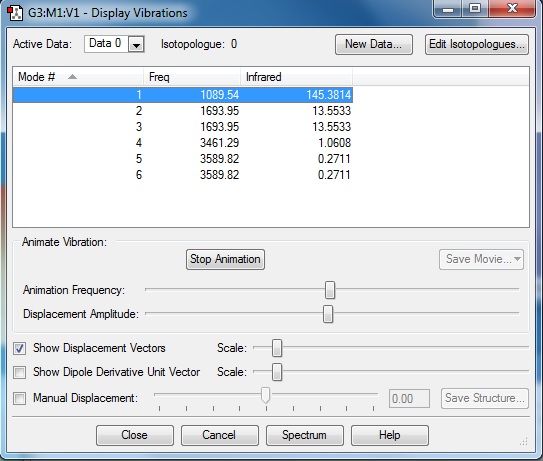
NH3 Display Vibrations
Questions
1. How many modes do you expect from the 3N-6 rule?
6
2. Which modes are degenerate (ie have the same energy)?
2,3 and 5,6
3. Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending: 1, 2, 3 & Stretching: 4, 5, 6
4. Which mode is highly symmetric?
4
5. One mode is known as the "umbrella" mode, which one is this?
1
6. How many bands would you expect to see in an experimental spectrum of gaseous ammonia?
2 Peaks will be seen in the IR spectra of gaseous ammonia because for a peak to occur there must be a change in the dipole moment. Modes 4, 5 and 6 do not have a change in dipole so will not be seen and 2&3 are degenerate will appear as a singular peak, therefore there will be a total of 2 peaks.
Atomic Charges - N has a charge of -1.125 - H has a charge of +0.375
This is what is expected as nitrogen is more electronegative than hydrogen so has a greater electron density.
N2 Molecule
Molecule Name : Nitrogen
Calculation Method : RB3LYP
Basis Set : 6-31G(d,p)
Final Energy E(RB3LYP) : -109.52412868 a.u.
RMS Gradient : 0.00000060 a.u.
Point Group : D*H
N-N Bond Distance : 1.10550
N-N Bond Angle : 180.000
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
NH molecule |
The optimisation file is linked here
N2 Display Vibrations
Questions
1. How many modes do you expect from the 3N-5 rule?
1
2. Which modes are degenerate (ie have the same energy)?
N/A
3. Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending: N/A Stretching: 1
4. Which mode is highly symmetric?
1
5. One mode is known as the "umbrella" mode, which one is this?
N/A
6. How many bands would you expect to see in an experimental spectrum of nitrogen?
No bands will be seen in the experimental spectra because there will be no change in the dipole moment of the molecule so it is not IR active hence no bands will be seen.
Atomic Charges - Both N atoms have a charge of 0.0
H2 Molecule
Molecule Name : Hydrogen
Calculation Method : RB3LYP
Basis Set : 6-31G(d,p)
Final Energy E(RB3LYP) : -1.17853936 a.u.
RMS Gradient : 0.00000017 a.u.
Point Group : D*H
H-H Bond Distance : 0.74279
H-H Bond Angle : 180.000
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
NH molecule |
The optimisation file is linked here
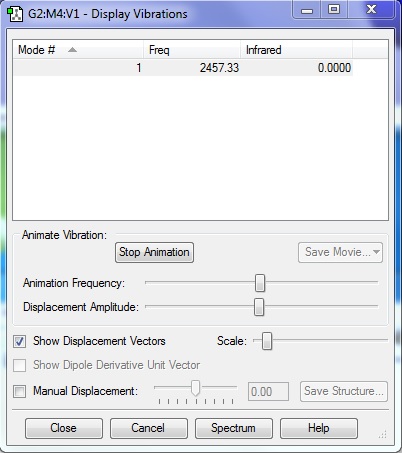
H2 Display Vibrations
Questions
1. How many modes do you expect from the 3N-5 rule?
1
2. Which modes are degenerate (ie have the same energy)?
N/A
3. Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending: N/A Stretching: 1
4. Which mode is highly symmetric?
1
5. One mode is known as the "umbrella" mode, which one is this?
N/A
6. How many bands would you expect to see in an experimental spectrum of hydrogen?
No bands will be seen in the experimental spectra because there will be no change in the dipole moment of the molecule so it is not IR active hence no bands will be seen.
Atomic Charges - Both H atoms have a charge of 0.0
Reaction Energies
E(NH3)= -56.55776873 a.u. 2*E(NH3)= -113.1155375 a.u. E(N2)= -109.52412868 a.u. E(H2)= -1.17853936 a.u. 3*E(H2)= -3.53561808 a.u. ΔE=2*E(NH3)-[E(N2)+3*E(H2)]= -0.0557907 a.u.
-0.0557907 a.u. x 2625.5 = -146.4784829 kJ/mol
Explanation
This is an exothermic reaction therefore, the reactants are at a higher energy than the product. As a result, the product are more stable as they are lower in energy.
Literature The value in the literature is -92.2[1] which is higher than the value that has been calculated above. This difference could be a cause of poor optimization and this is a theoretical value.
Project Molecule - HCl
Molecule Name : Hydrogen Chloride
Calculation Method : RB3LYP
Basis Set : 6-31G(d,p)
Final Energy E(RB3LYP) : -460.80077875 a.u.
RMS Gradient : 0.00005211 a.u.
Point Group : C*V
H-Cl Bond Distance : 1.28599
H-Cl Bond Angle : 180.000
Item Value Threshold Converged?
Maximum Force 0.000090 0.000450 YES
RMS Force 0.000090 0.000300 YES
Maximum Displacement 0.000139 0.001800 YES
RMS Displacement 0.000197 0.001200 YES
HCl molecule |
The optimisation file is linked here
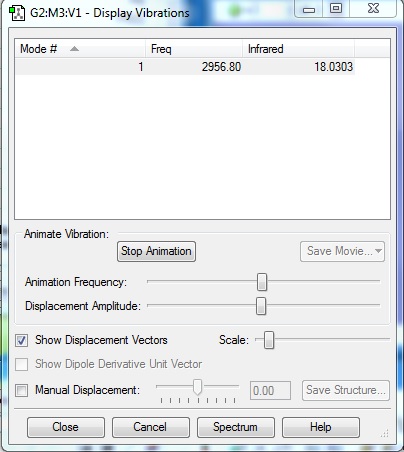
HCl Display Vibrations
Questions
1. How many modes do you expect from the 3N-5 rule?
1
2. Which modes are degenerate (ie have the same energy)?
N/A
3. Which modes are "bending" vibrations and which are "bond stretch" vibrations?
Bending: N/A Stretching: 1
4. Which mode is highly symmetric?
1
5. One mode is known as the "umbrella" mode, which one is this?
N/A
6. How many bands would you expect to see in an experimental spectrum of hydrogen chloride?
1 band will be seen in the experimental spectrum for HCl because there is one asymmetric stretch that can be seen. Additionally, the difference in electronegativities means that there is a change in dipole moment hence this molecule is microwave active.
Atomic Charges - H has a charge of +0.284 - Cl has a charge of -0.284
This is expected as Cl has an electronegativity of 3.16 whilst H has an electronegativity of 2.20. Therefore, there will be a greater distribution of electron density around chlorine thus a negative charge is seen on Cl.
Molecular Orbitals
This is a molecular orbital that has an energy level that is just below that of the HOMO_LUMO region, this means that this is the HOMO. This is going to have a large influence on the bond character as these are valence electrons. It is showing the p orbitals on Cl and is spread across the entirety of the bond.
This molecular orbital is depicting the atomic orbital of a 2p orbital, distinguished by the node. This p orbital is orthogonal to the bond. This is quite deep in energy and is occupied by 2 electrons. It is unlikely that this orbital will contribute to bonding as it is not in the right alignment to overlap with molecular orbitals on the H atom.
Similarly, to the molecular orbital image above this is representing a 2s orbital of chlorine.It is not involved in bonding as it is not interacting with any other orbitals. This is deeper in energy than the p orbitals as it is closer to the nucleus, therefore it is further away from the HOMO-LUMO range.
This image is an atomic orbital and another orientation of the second image. It shows the 2p orbital of chlorine, recognized by the distinctive node. As seen above this orbital is not interacting with anything else so is not impacting the bonding. This is deeper in energy as it is more negative and is also degenerate, has the same energy level as image 2.
This image is depicting a molecular orbital between two s orbitals from H and Cl. This MO is a bonding orbital and it is high in energy as it approaches the HOMO-LUMO range and is occupied by two electrons. As this is high in energy it is likely to have a large effect on bonding.
References
[1]Periodicity, Quantitative Equilibrium and Functional Group Chemistry Nelson Advanced Science Series