Rep:Mod:01338675
Issy's Wiki Page
NH3
Information on NH3 Molecule
| Identity | NH3 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final energy E(RB3LYP) (au) | -56.55776873 |
| RMS Gradient (au) | 0.00000485 |
| Point Group | C3V |
| Optimised N-H Bond Length (Å) | 1.01798 |
| Optimised H-N-H Bond Angle | 105.741 |
The literature value for bond length is 1.008 Å [1] which is reasonably close to the calculated value of 1.01798 Å. The difference may be attributed to the rounding errors of the computer.
'Item' Table for Optimised NH3 Molecule
Item Value Threshold Converged?
Maximum Force 0.000004 0.000450 YES
RMS Force 0.000004 0.000300 YES
Maximum Displacement 0.000072 0.001800 YES
RMS Displacement 0.000035 0.001200 YES
Predicted change in Energy=-5.986294D-10
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.018 -DE/DX = 0.0 !
! R2 R(1,3) 1.018 -DE/DX = 0.0 !
! R3 R(1,4) 1.018 -DE/DX = 0.0 !
! A1 A(2,1,3) 105.7412 -DE/DX = 0.0 !
! A2 A(2,1,4) 105.7412 -DE/DX = 0.0 !
! A3 A(3,1,4) 105.7412 -DE/DX = 0.0 !
! D1 D(2,1,4,3) -111.8571 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
JMol Image
test molecule |
The optimisation file is liked to here
NH3 Vibrations

| Expected Number of Modes | 6 |
| Number of Degenerate Modes | 4 |
| Identity of Degenerate Modes | 2 and 3; 5 and 6 |
| Identity of Bending Modes | 1, 2, 3 |
| Identity of Stretching Modes | 4, 5, 6 |
| Highly Symmetric Mode | 1 (Bending) and 4 (Stretching) |
| 'Umbrella Mode' | 1 |
| Number of Bands Expected in Spectrum | 2 |
The expected number of modes was calculated using the equation: number of modes = 3N-6, which is valid for non-linear molecules like ammonia. The number and identity of degenerate modes were identified by looking at the values for the modes. 2 and 3 had the same frequency, as did 5 and 6 meaning altogether there were 4 degenerate modes. The identity of the bending and stretching modes were decided by looking at the animations. The highly symmetric modes and the umbrella mode were identified using the animations also. The number of bands expected would be 2 despite the fact there are modes at 4 different energies. The intensity of the stretching peaks is so small that they don't appear in the spectrum as the change in dipole moment is so small that among the noise of the spectrum it would be imperceptible.
NH3 Charge
| Atom | Charge |
|---|---|
| N | -1.125 |
| H | 0.375 |
I would expect the nitrogen atom to carry a negative charge as it is more electronegative than hydrogen. These numbers are sensible because the overall charge is 0 and ammonia is a neutral molecule.
N2
Information on N2
| Identity | N2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final energy E(RB3LYP) (au) | -109.52412868 |
| RMS Gradient (au) | 0.00000060 |
| Point Group | D∞h |
| Optimised N-N Bond Length (Å) | 1.10550 |
| Optimised N-N Bond Angle | 180 |
The literature value for bond length is 1.0975 Å [1] which is reasonably close to the calculated value of 1.10550 Å. The difference may be attributed to the rounding errors of the computer.
'Item' Table for Optimised N2 Molecule
Item Value Threshold Converged?
Maximum Force 0.000001 0.000450 YES
RMS Force 0.000001 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000000 0.001200 YES
Predicted change in Energy=-3.400980D-13
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 1.1055 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
N2 Vibrations
There is a stretching mode at 2457.33 cm-1. This mode is a symmetrical stretch so there is no change in dipole moment. This means it would not be an IR active stretch.
JMol Image
test molecule |
The optimisation file is liked to here
MOs of N2
H2
Information on H2
| Identity | H2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final energy E(RB3LYP) (au) | -1.17853936 |
| RMS Gradient (au) | 0.00000017 |
| Point Group | D∞h |
| Optimised H-H Bond Length (Å) | 0.74279 |
| Optimised H-H Bond Angle | 180 |
The literature value for bond length is 74 Å [2] which is reasonably close to the calculated value of 0.74279 Å. The difference may be attributed to the rounding errors of the computer.
'Item' Table for Optimised H2 Molecule
Item Value Threshold Converged?
Maximum Force 0.000000 0.000450 YES
RMS Force 0.000000 0.000300 YES
Maximum Displacement 0.000000 0.001800 YES
RMS Displacement 0.000001 0.001200 YES
Predicted change in Energy=-1.164080D-13
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 0.7428 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
H2 Vibrations
There is a stretching mode at 4465.58 cm-1. This mode is a symmetrical stretch so there is no change in dipole moment. This means it would not be an IR active stretch.
JMol Image
test molecule |
The optimisation file is liked to here
Energies
| E(NH3) | -56.55776873 |
| 2*E(NH3) | -113.11553746 |
| E(N2) | -109.52412868 |
| E(H2) | -1.17853936 |
| 3*E(H2) | -3.53561808 |
| ΔE=2*E(NH3)-[E(N2)+3*E(H2)] | -146.47848285 (kJmol-1) |
The energy change reaction is -146.5 kJmol-1 meaning energy is released, so it is an exothermic reaction. The exothermic nature of this reaction means that the ammonia product is more stable than the gaseous reactants. This energy change equation works because it uses the E(reaction) = E(products)-E(reactants). Some values of energy are multiplied by either 2 or 3 which reflects the stoichiometry of the reaction (N2 + 3H2 --> 2NH3).
Own Molecule: Cl2
Information on Cl2
| Identity | Cl2 |
| Calculation Method | RB3LYP |
| Basis Set | 6-31G(d,p) |
| Final energy E(RB3LYP) (au) | -920.34987886 |
| RMS Gradient (au) | 0.00002510 |
| Point Group | D∞h |
| Optimised Cl-Cl Bond Length (Å) | 2.04174 |
| Optimised Cl-Cl Bond Angle | 180 |
The literature value for bond length is 1.99 Å [3] which is reasonably close to the calculated value of 2.04174 Å. The difference may be attributed to the rounding errors of the computer.
'Item' Table for Optimised Cl2 Molecule
Item Value Threshold Converged?
Maximum Force 0.000043 0.000450 YES
RMS Force 0.000043 0.000300 YES
Maximum Displacement 0.000121 0.001800 YES
RMS Displacement 0.000172 0.001200 YES
Predicted change in Energy=-5.277110D-09
----------------------------
! Optimized Parameters !
! (Angstroms and Degrees) !
-------------------------- --------------------------
! Name Definition Value Derivative Info. !
--------------------------------------------------------------------------------
! R1 R(1,2) 2.0417 -DE/DX = 0.0 !
--------------------------------------------------------------------------------
Cl2 Vibrations
There is a stretching mode at 520.32 cm-1. This mode is a symmetrical stretch so there is no change in dipole moment. This means it would not be an IR active stretch.
JMol Image
test molecule |
The optimisation file is liked to here
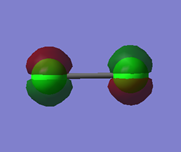
Charge on Cl2

The image shows the charge distribution on the Cl2 molecule. This is as expected because Cl2 is a neutral diatomic molecule so the electronegativies of the atoms are identical so there is no overall charge.
MOs for Cl2
The non-bonding, bonding and anti-bonding MOs were identified by looking at whether a large cloud of electron density covered the molecule and whether there was a node at the middle of the bond. There is no spherical cloud of electron density that surrounds the whole molecule in non-bonding molecular orbitals and a node is present in anti-bonding molecular orbitals. All electrons are paired in Cl2 meaning it is diamagnetic.
References
- ↑ 1.0 1.1 Tables of Interatomic Distances and Configuration in Molecules and Ions, L.E. Sutton, ed., London: The Chemical Society, 1958
- ↑ Common Bond Energies, Wired Chemist: http://www.wiredchemist.com/chemistry/data/bond_energies_lengths.html
- ↑ Bond Lengths and Energies, Wired Chemist: http://www.science.uwaterloo.ca/~cchieh/cact/c120/bondel.html